%load_ext autoreload
%autoreload 2
Restructuring PubMed Abstracts using NLP¶
Milestone Project 2¶
In the previous notebook, we learned about tokenization (breaking up text into tokens) and creating embeddings (learning a numeric representation of the tokens)
In this notebook, we are going to implement the deep learning model from 2017 paper PubMed 200k RCT: a Dataset for Sequenctial Sentence Classification in Medical Abstracts.
- The paper presented a new dataset called PubMed 200k RCT which consists of ~200K labelled Randomized Controlled Trial abstracts.
- Goal is to predict sentences which follow sequentially into broadly 4 categories
OBJECTIVE,METHODS,RESULTS,CONCLUSIONS

- Given the abstract of the research paper, which category does each sentence fall into?
Model Input¶
Following abstract input (digits replaced with @ symbol)
To investigate the efficacy of @ weeks of daily low-dose oral prednisolone in improving pain , mobility , and systemic low-grade inflammation in the short term and whether the effect would be sustained at @ weeks in older adults with moderate to severe knee osteoarthritis ( OA ). A total of @ patients with primary knee OA were randomized @:@ ; @ received @ mg/day of prednisolone and @ received placebo for @ weeks. Outcome measures included pain reduction and improvement in function scores and systemic inflammation markers. Pain was assessed using the visual analog pain scale ( @-@ mm ). Secondary outcome measures included the Western Ontario and McMaster Universities Osteoarthritis Index scores , patient global assessment ( PGA ) of the severity of knee OA , and @-min walk distance ( @MWD )., Serum levels of interleukin @ ( IL-@ ) , IL-@ , tumor necrosis factor ( TNF ) - , and high-sensitivity C-reactive protein ( hsCRP ) were measured. There was a clinically relevant reduction in the intervention group compared to the placebo group for knee pain , physical function , PGA , and @MWD at @ weeks. The mean difference between treatment arms ( @ % CI ) was @ ( @-@ @ ) , p < @ ; @ ( @-@ @ ) , p < @ ; @ ( @-@ @ ) , p < @ ; and @ ( @-@ @ ) , p < @ , respectively. Further , there was a clinically relevant reduction in the serum levels of IL-@ , IL-@ , TNF - , and hsCRP at @ weeks in the intervention group when compared to the placebo group. These differences remained significant at @ weeks. The Outcome Measures in Rheumatology Clinical Trials-Osteoarthritis Research Society International responder rate was @ % in the intervention group and @ % in the placebo group ( p < @ ). Low-dose oral prednisolone had both a short-term and a longer sustained effect resulting in less knee pain , better physical function , and attenuation of systemic inflammation in older patients with knee OA ( ClinicalTrials.gov identifier NCT@ ).
Model Output¶
Separate out the sentences and mark the category
['###24293578\n', \ 'OBJECTIVE\tTo investigate the efficacy of @ weeks of daily low-dose oral prednisolone in improving pain , mobility , and systemic low-grade inflammation in the short term and whether the effect would be sustained at @ weeks in older adults with moderate to severe knee osteoarthritis ( OA ) .\n', \ 'METHODS\tA total of @ patients with primary knee OA were randomized @:@ ; @ received @ mg/day of prednisolone and @ received placebo for @ weeks .\n', 'METHODS\tOutcome measures included pain reduction and improvement in function scores and systemic inflammation markers .\n', \ 'METHODS\tPain was assessed using the visual analog pain scale ( @-@ mm ) .\n', 'METHODS\tSecondary outcome measures included the Western Ontario and McMaster Universities Osteoarthritis Index scores , patient global assessment ( PGA ) of the severity of knee OA , and @-min walk distance ( @MWD ) .\n', \ 'METHODS\tSerum levels of interleukin @ ( IL-@ ) , IL-@ , tumor necrosis factor ( TNF ) - , and high-sensitivity C-reactive protein ( hsCRP ) were measured .\n', \ 'RESULTS\tThere was a clinically relevant reduction in the intervention group compared to the placebo group for knee pain , physical function , PGA , and @MWD at @ weeks .\n', \ 'RESULTS\tThe mean difference between treatment arms ( @ % CI ) was @ ( @-@ @ ) , p < @ ; @ ( @-@ @ ) , p < @ ; @ ( @-@ @ ) , p < @ ; and @ ( @-@ @ ) , p < @ , respectively .\n', \ 'RESULTS\tFurther , there was a clinically relevant reduction in the serum levels of IL-@ , IL-@ , TNF - , and hsCRP at @ weeks in the intervention group when compared to the placebo group .\n', 'RESULTS\tThese differences remained significant at @ weeks .\n', 'RESULTS\tThe Outcome Measures in Rheumatology Clinical Trials-Osteoarthritis Research Society International responder rate was @ % in the intervention group and @ % in the placebo group ( p < @ ) .\n', \ 'CONCLUSIONS\tLow-dose oral prednisolone had both a short-term and a longer sustained effect resulting in less knee pain , better physical function , and attenuation of systemic inflammation in older patients with knee OA ( ClinicalTrials.gov identifier NCT@ ) .\n', \ '\n']
Problem with unstructured abstracts¶
Papers without structured abstracts can be hard to read.
Solution - Make it structured¶
Build a NLP model to classify each sentence into the appropriate category -> allow researchers to skim through the literature with ease.
Create an NLP model to classify abstract sentences into the role they play (e.g. objective, methods, results, etc) to enable researchers to skim through the literature (hence SkimLit 🤓🔥) and dive deeper when necessary.
Resource:
Contents of this notebook¶
- Download the text dataset
- Preprocess the dataset into appropriate structure for modelling
- Set up a series of modelling experiments:
- Naive Bayes (TF-IDF) baseline
- Deep models with different combinations of: token embeddings, character embeddings, pretrained embeddings, positional embeddings
Build a multimodel model (which takes more than one input)
- Replicate the architecture from Neural networks for joint sentence classification in medical paper abstracts.
Find the model's most wrong prediction
- Make predictions on Pubmed abstracts from the internet
Change to project directory¶
import os
os.chdir('/content/drive/MyDrive/projects/Tensorflow-tutorial-Daniel-Bourke/notebooks')
import sys
sys.path.append('../')
Access to GPU¶
!nvidia-smi -L
NVIDIA-SMI has failed because it couldn't communicate with the NVIDIA driver. Make sure that the latest NVIDIA driver is installed and running.
Download the data¶
There are four versions of the dataset
Pubmed 200kPubmed 200k with at sign- Here digits are replaced with @ symbolPubmed 20kPubmed 20k with at sign
The README.md file has the following information:
- PubMed 20k is a subset of PubMed 200k. I.e., any abstract present in PubMed 20k is also present in PubMed 200k.
PubMed_200k_RCTis the same asPubMed_200k_RCT_numbers_replaced_with_at_sign, except that in the latter all numbers had been replaced by@. (same forPubMed_20k_RCTvs.PubMed_20k_RCT_numbers_replaced_with_at_sign).- Since Github file size limit is 100 MiB, we had to compress
PubMed_200k_RCT\train.7zandPubMed_200k_RCT_numbers_replaced_with_at_sign\train.zip. To uncompresstrain.7z, you may use 7-Zip on Windows, Keka on Mac OS X, or p7zip on Linux.
# !git clone https://github.com/Franck-Dernoncourt/pubmed-rct.git /content/data
# !mv /content/data/* ../data/
To make our training faster we will experiment with the 20k version of the dataset.
!ls ../data/PubMed_20k_RCT_numbers_replaced_with_at_sign/
dev.txt test.txt train.txt
There are three sets train, test and dev
data_dir = '../data/PubMed_20k_RCT_numbers_replaced_with_at_sign/'
import os
filenames = os.listdir(data_dir)
filenames
['train.txt', 'dev.txt', 'test.txt']
filenames = {f.replace('.txt', ''): os.path.join(data_dir, f) for f in filenames}
filenames
{'dev': '../data/PubMed_20k_RCT_numbers_replaced_with_at_sign/dev.txt',
'test': '../data/PubMed_20k_RCT_numbers_replaced_with_at_sign/test.txt',
'train': '../data/PubMed_20k_RCT_numbers_replaced_with_at_sign/train.txt'}
Great now we have parsed the filenames, let us preprocess the data
Preprocess the data¶
def read_lines(fname):
with open(fname, mode='r') as f:
return f.readlines()
train_lines = read_lines(filenames['train'])
train_lines[:20]
['###24293578\n', 'OBJECTIVE\tTo investigate the efficacy of @ weeks of daily low-dose oral prednisolone in improving pain , mobility , and systemic low-grade inflammation in the short term and whether the effect would be sustained at @ weeks in older adults with moderate to severe knee osteoarthritis ( OA ) .\n', 'METHODS\tA total of @ patients with primary knee OA were randomized @:@ ; @ received @ mg/day of prednisolone and @ received placebo for @ weeks .\n', 'METHODS\tOutcome measures included pain reduction and improvement in function scores and systemic inflammation markers .\n', 'METHODS\tPain was assessed using the visual analog pain scale ( @-@ mm ) .\n', 'METHODS\tSecondary outcome measures included the Western Ontario and McMaster Universities Osteoarthritis Index scores , patient global assessment ( PGA ) of the severity of knee OA , and @-min walk distance ( @MWD ) .\n', 'METHODS\tSerum levels of interleukin @ ( IL-@ ) , IL-@ , tumor necrosis factor ( TNF ) - , and high-sensitivity C-reactive protein ( hsCRP ) were measured .\n', 'RESULTS\tThere was a clinically relevant reduction in the intervention group compared to the placebo group for knee pain , physical function , PGA , and @MWD at @ weeks .\n', 'RESULTS\tThe mean difference between treatment arms ( @ % CI ) was @ ( @-@ @ ) , p < @ ; @ ( @-@ @ ) , p < @ ; @ ( @-@ @ ) , p < @ ; and @ ( @-@ @ ) , p < @ , respectively .\n', 'RESULTS\tFurther , there was a clinically relevant reduction in the serum levels of IL-@ , IL-@ , TNF - , and hsCRP at @ weeks in the intervention group when compared to the placebo group .\n', 'RESULTS\tThese differences remained significant at @ weeks .\n', 'RESULTS\tThe Outcome Measures in Rheumatology Clinical Trials-Osteoarthritis Research Society International responder rate was @ % in the intervention group and @ % in the placebo group ( p < @ ) .\n', 'CONCLUSIONS\tLow-dose oral prednisolone had both a short-term and a longer sustained effect resulting in less knee pain , better physical function , and attenuation of systemic inflammation in older patients with knee OA ( ClinicalTrials.gov identifier NCT@ ) .\n', '\n', '###24854809\n', 'BACKGROUND\tEmotional eating is associated with overeating and the development of obesity .\n', 'BACKGROUND\tYet , empirical evidence for individual ( trait ) differences in emotional eating and cognitive mechanisms that contribute to eating during sad mood remain equivocal .\n', 'OBJECTIVE\tThe aim of this study was to test if attention bias for food moderates the effect of self-reported emotional eating during sad mood ( vs neutral mood ) on actual food intake .\n', 'OBJECTIVE\tIt was expected that emotional eating is predictive of elevated attention for food and higher food intake after an experimentally induced sad mood and that attentional maintenance on food predicts food intake during a sad versus a neutral mood .\n', 'METHODS\tParticipants ( N = @ ) were randomly assigned to one of the two experimental mood induction conditions ( sad/neutral ) .\n']
The format of the dataset is as follows:
- Starting of every new abstract is indicated by three hash symbols
#followed by the abstract id - The category of the sentence is marked as
<CATEGORY_NAME>\t - Finally, the newline symbol
\nmarks the end of the abstract
Question: How should we structure our abstract into a parseable information?
For each abstract we can return a dictionary which we keep appending to a list as follows:
[{'line_number': 0, 'target': 'OBJECTIVE', 'text': 'to investigate the efficacy of @ weeks of daily low-dose oral prednisolone in improving pain , mobility , and systemic low-grade inflammation in the short term and whether the effect would be sustained at @ weeks in older adults with moderate to severe knee osteoarthritis ( oa ) .', 'total_lines': 11}, ...]
def parse_structure_of_abstract_file(fpath):
lines = read_lines(fpath)
abstract_samples = []
line_number = 0
total_lines = 0
for line in lines:
if line.startswith('###'):
abstract_id = line.replace('###', '').replace('\n', '')
abstract_lines = ''
elif line.isspace():
abstract_lines_lst = abstract_lines.splitlines()
total_lines = len(abstract_lines_lst) - 1
for line_number, abstract_line in enumerate(abstract_lines_lst):
line_data = {}
line_data['id'] = abstract_id
line_data['target'], line_data['text'] = abstract_line.split('\t')
line_data['line_number'] = line_number
line_data['total_lines'] = total_lines
abstract_samples.append(line_data)
else:
abstract_lines += line
return abstract_samples
data_samples = {}
for subset, filename in filenames.items():
data_samples[subset] = parse_structure_of_abstract_file(filename)
data_samples['train'][:5]
[{'id': '24293578',
'line_number': 0,
'target': 'OBJECTIVE',
'text': 'To investigate the efficacy of @ weeks of daily low-dose oral prednisolone in improving pain , mobility , and systemic low-grade inflammation in the short term and whether the effect would be sustained at @ weeks in older adults with moderate to severe knee osteoarthritis ( OA ) .',
'total_lines': 11},
{'id': '24293578',
'line_number': 1,
'target': 'METHODS',
'text': 'A total of @ patients with primary knee OA were randomized @:@ ; @ received @ mg/day of prednisolone and @ received placebo for @ weeks .',
'total_lines': 11},
{'id': '24293578',
'line_number': 2,
'target': 'METHODS',
'text': 'Outcome measures included pain reduction and improvement in function scores and systemic inflammation markers .',
'total_lines': 11},
{'id': '24293578',
'line_number': 3,
'target': 'METHODS',
'text': 'Pain was assessed using the visual analog pain scale ( @-@ mm ) .',
'total_lines': 11},
{'id': '24293578',
'line_number': 4,
'target': 'METHODS',
'text': 'Secondary outcome measures included the Western Ontario and McMaster Universities Osteoarthritis Index scores , patient global assessment ( PGA ) of the severity of knee OA , and @-min walk distance ( @MWD ) .',
'total_lines': 11}]
import pandas as pd
data_samples = {subset: pd.DataFrame(data) for subset, data in data_samples.items()}
data_samples['train'].head()
| id | target | text | line_number | total_lines | |
|---|---|---|---|---|---|
| 0 | 24293578 | OBJECTIVE | To investigate the efficacy of @ weeks of dail... | 0 | 11 |
| 1 | 24293578 | METHODS | A total of @ patients with primary knee OA wer... | 1 | 11 |
| 2 | 24293578 | METHODS | Outcome measures included pain reduction and i... | 2 | 11 |
| 3 | 24293578 | METHODS | Pain was assessed using the visual analog pain... | 3 | 11 |
| 4 | 24293578 | METHODS | Secondary outcome measures included the Wester... | 4 | 11 |
CLASS_NAMES = sorted(list(data_samples['train']['target'].unique()))
CLASS_NAMES
['BACKGROUND', 'CONCLUSIONS', 'METHODS', 'OBJECTIVE', 'RESULTS']
N_CLASSES = len(CLASS_NAMES)
# Let's see the distribution of target categories
data_samples['train'].target.value_counts()
METHODS 59353 RESULTS 57953 CONCLUSIONS 27168 BACKGROUND 21727 OBJECTIVE 13839 Name: target, dtype: int64
data_samples['train'].total_lines.value_counts().sort_index().plot(kind='bar');
Most of the abstracts are around 7 to 15 lines long, with the modal value at 11.
Get lists of sentences¶
When we train our deep neural networks, depending upon how we build the ingestion layer/input layer, we might have to pass the input sentences as a list.
sentences = {subset: data['text'].tolist() for subset, data in data_samples.items()}
Make numeric labels (ML models require numeric labels)¶
We will create both one-hot-encoded labels and integer labels.
data_labels = {'one_hot': {}, 'label': {}}
One Hot Encode¶
from sklearn.preprocessing import OneHotEncoder, LabelEncoder
one_hot_encoder = OneHotEncoder(sparse=False)
data_labels['one_hot']['train'] = one_hot_encoder.fit_transform(data_samples['train'].target.to_numpy().reshape(-1, 1))
for subset in ['dev', 'test']:
data_labels['one_hot'][subset] = one_hot_encoder.transform(data_samples[subset].target.to_numpy().reshape(-1, 1))
Label Encode¶
label_encoder = LabelEncoder()
data_labels['label']['train'] = label_encoder.fit_transform(data_samples['train'].target.to_numpy())
for subset in ['dev', 'test']:
data_labels['label'][subset] = label_encoder.transform(data_samples[subset].target.to_numpy())
data_labels['one_hot']['train']
array([[0., 0., 0., 1., 0.],
[0., 0., 1., 0., 0.],
[0., 0., 1., 0., 0.],
...,
[0., 0., 0., 0., 1.],
[0., 1., 0., 0., 0.],
[0., 1., 0., 0., 0.]])
data_labels['label']['train']
array([3, 2, 2, ..., 4, 1, 1])
Create a series of modelling experiments¶
We will proceed with our modelling process as follows:
- Start with the most basic Multinomial Naive Bayes model with TF-IDF
- Then we will build more and more complex deep learning models
- Finally we will lead it up to implementing the architecture in the paper Neural networks for joint sentence classification in medical paper abstracts.
MODELS = {}
PREDICTIONS = {}
Model 0: Naive Bayes Baseline¶
model_name = 'naive-bayes-baseline'
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.naive_bayes import MultinomialNB
from sklearn.pipeline import Pipeline
model = Pipeline([
('vectorize', TfidfVectorizer()),
('clf', MultinomialNB())
])
model.fit(sentences['train'], data_labels['label']['train'])
MODELS[model_name] = model
model.score(sentences['dev'], data_labels['label']['dev'])
0.7218323844829869
Now we have to first target to beat atleast this 72.1 % accuracy of the baseline model.
from src.utils import reshape_classification_prediction
# Make predictions
PREDICTIONS[model_name] = {}
for subset, sents in sentences.items():
PREDICTIONS[model_name][subset] = reshape_classification_prediction(model.predict_proba(sents))
from sklearn.metrics import classification_report
print(classification_report(PREDICTIONS[model_name]['dev'].argmax(axis=1), data_labels['label']['dev'], target_names=CLASS_NAMES))
precision recall f1-score support
OBJECTIVE 0.49 0.66 0.56 2568
METHODS 0.59 0.65 0.61 4159
RESULTS 0.87 0.72 0.79 11970
CONCLUSIONS 0.14 0.75 0.23 435
BACKGROUND 0.86 0.76 0.81 11080
accuracy 0.72 30212
macro avg 0.59 0.71 0.60 30212
weighted avg 0.78 0.72 0.74 30212
Preparing our data for deep sequence models¶
- Before we start training our deep neural nets we need to text vectorization and embedding layers before the Input layer of the model.
import matplotlib.pyplot as plt
sent_lens = [len(sent.split()) for sent in sentences['train']]
plt.hist(sent_lens);
Majority of the sentences are between 0 and 50 tokens long. Let us limit the maximum sequence length to cover 95% of the samples (this can possibly prevent overfitting)
import numpy as np
max_seq_length = int(np.percentile(sent_lens, 95))
max_seq_length
55
Note: From reading the section 4 of the PubMed 200k RCT paper paper, it is a good idea to look at the length distribution of the sentences in the corpus.
Create text vectorizer¶
- Tokenizing sentence, and converting text into integers (each token mapped to a unique integer in the vocabulary)
- Section 3.2 of the PubMed 200k RCT paper paper limits the maximum vocabulary size to 68000
max_tokens = 68000
from tensorflow.keras.layers.experimental.preprocessing import TextVectorization
text_vectorizer = TextVectorization(max_tokens=max_tokens,
standardize='lower_and_strip_punctuation',
split='whitespace',
output_sequence_length=max_seq_length)
text_vectorizer.adapt(sentences['train'])
# Let's check on a random sentence
rand_sent = np.random.choice(sentences['dev'])
sent_vec = text_vectorizer([rand_sent])
total_tokens = len(sent_vec[0])
zero_pads = (sent_vec[0] == 0).numpy().sum()
actual_tokens = total_tokens - zero_pads
print('Text:\n', rand_sent, '\n')
print(f'Number of tokens: total={total_tokens}, actual={actual_tokens}, zero_pad={zero_pads}\n')
print('Vectorized text:\n', sent_vec)
Text:
Therefore , it failed to answer the question as to whether insomnia is , indeed , a risk factor for increased headache frequency and headache intensity in migraineurs .
Number of tokens: total=55, actual=25, zero_pad=30
Vectorized text:
tf.Tensor(
[[ 709 185 1297 6 5527 2 2395 25 6 180 1591 20
11144 8 73 432 11 96 1309 400 3 1309 579 5
10260 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0]], shape=(1, 55), dtype=int64)
Let us get the vocabulary of the training corpus using the get_vocabulary() method of the TextVectorizer
vocab = text_vectorizer.get_vocabulary()
print('Number of tokens in the vocabulary:', len(vocab))
print('Most common top 5 tokens:', vocab[:5])
print('Least common bottom 5 tokens:', vocab[-5:])
Number of tokens in the vocabulary: 64841 Most common top 5 tokens: ['', '[UNK]', 'the', 'and', 'of'] Least common bottom 5 tokens: ['aainduced', 'aaigroup', 'aachener', 'aachen', 'aaacp']
The configuration of the TextVectorizer
text_vectorizer.get_config()
{'dtype': 'string',
'max_tokens': 68000,
'name': 'text_vectorization_1',
'ngrams': None,
'output_mode': 'int',
'output_sequence_length': 55,
'pad_to_max_tokens': False,
'split': 'whitespace',
'standardize': 'lower_and_strip_punctuation',
'trainable': True,
'vocabulary_size': 64841}
Create custom text embedding¶
- Our
text_vextorizermaps the tokens to a unique integer id. But that is not a very meaningful representation of our tokens. - A numerical representation must be somewhat related to the actual meaning of the word (semantic meaning), may be context aware (hence sentence embeddings rather than word embeddings).
- We can use a pretrained embedding which encodes the knowledge of the words it has learned on a large text corpus (such as wikipedia). We can also fine-tune it for our specific dataset.
- Or we can learn a new embedding altogether, i.e. decide that we want a
d-dimensional numerical representation for each of our word, and adjust its weights by learning from the task. - The input dimension
input_dimto the embedding layer would be the size of the vocabulary and theoutput_dimis the one we decide i.e. 128 or 256 or whatever.
Here is how the embedding layer works:
- It randomly initializes a matrix of size
(num_vocab, embed_dim)by drawing from any probability distribution, say uniform. - Now for each word, it is simply a lookup for each token_id (assigned from the
text_vectorizer), by indexing into the embedding matrixembed_mat[idx]or by a simple matrix multiplication with a one-hot encoded vector. - So for each token we get a
ddimensional feature vector. Now we if we settrainable=True, these can be treated as weights in the gradient descent steps, and hence we can learn ad-dimensional encoded representation best suited for our dataset
from tensorflow.keras import layers
word_embed = layers.Embedding(input_dim=len(vocab), output_dim=128,
mask_zero=True,
name='word_embed')
Let's try out an example
rand_sent = np.random.choice(sentences['train'])
sent_vec = text_vectorizer([rand_sent])
sent_embed = word_embed(sent_vec)
print(f'Text:\n{rand_sent}\n')
print(f'Tokenized Shape={sent_vec.shape}:\n{sent_vec}\n')
print(f'Embedded Shape={sent_embed.shape}:\n{sent_embed}\n')
Text:
To overcome this problem , different acquisition techniques have been proposed , including the computed tomographic-based attenuation correction method .
Tokenized Shape=(1, 55):
[[ 6 4573 23 1339 197 2601 824 99 167 1820 251 2
1490 43532 3271 2055 363 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0]]
Embedded Shape=(1, 55, 128):
[[[ 0.04858262 0.04572786 0.00618804 ... -0.02906777 -0.01279309
0.00665309]
[-0.01555011 -0.04458395 -0.02943052 ... -0.00530935 0.03543044
-0.0440763 ]
[-0.03862621 0.04873857 0.02948517 ... -0.01633852 0.01009755
0.00309818]
...
[-0.00143504 0.04715837 0.00742704 ... -0.04679319 0.02952261
0.00851489]
[-0.00143504 0.04715837 0.00742704 ... -0.04679319 0.02952261
0.00851489]
[-0.00143504 0.04715837 0.00742704 ... -0.04679319 0.02952261
0.00851489]]]
Fetching, preprocessing Datasets for model input (As efficient and fast as possible)
We will use the tf.Data API which allows faster data loading:
Since we already have sentences and labels for each subset, we will create a TensorSliceDataset first, then map a function (if we want to), batch it (batch_size=32), and then prefetch it (set tf.data.AUTOTUNE)
import tensorflow as tf
First make the TensorSliceDataset for each subset
tfdatasets = {'sent': {}, 'char': {}}
for subset in ['train', 'dev', 'test']:
tfdatasets['sent'][subset] = tf.data.Dataset.from_tensor_slices((sentences[subset], data_labels['one_hot'][subset]))
tfdatasets['sent']['train']
<TensorSliceDataset shapes: ((), (5,)), types: (tf.string, tf.float64)>
Now, batch and prefetch
for subset in ['train', 'dev', 'test']:
tfdatasets['sent'][subset] = tfdatasets['sent'][subset].batch(32).prefetch(tf.data.AUTOTUNE)
tfdatasets['sent']['train']
<PrefetchDataset shapes: ((None,), (None, 5)), types: (tf.string, tf.float64)>
NUM_EPOCHS = 50
Model 1: Conv1D with word embeddings¶
All of our deep models will follow a similar structure as below:
Input (text) -> tokenize -> embedding -> layers -> Output (softmax probability)
Here is where differences may occur:
- Input is already tokenized into integers
- Embedding maybe pretrained and we set the weights from
Word2Vec(say, trained on twitter corpus)- Keep it trainable or not
- Input may already be a series of embedding vectors (not recommended due to high size of vectors (maybe))
- Output may or may not be one-hot encoded
- One-hot encoded -> Use
categorical_crossentropyas the loss- Allows us to use
label_smoothing
- Allows us to use
- Label encoded (integers) -> Use
sparse_categorical_crossentropyas the loss
- One-hot encoded -> Use
from src.evaluate import KerasMetrics
from src.visualize import plot_learning_curve
model_name = 'Conv1D-word-embed'
inputs = layers.Input(shape=(1,), dtype=tf.string)
vectors = text_vectorizer(inputs)
embedding = word_embed(vectors)
x = layers.Conv1D(64, kernel_size=5, padding='same', activation='relu')(embedding)
x = layers.GlobalAveragePooling1D()(x)
outputs = layers.Dense(N_CLASSES, activation='softmax')(x)
model = tf.keras.models.Model(inputs, outputs, name=model_name)
model.compile(loss='categorical_crossentropy', optimizer=tf.keras.optimizers.Adam(),
metrics=['accuracy', KerasMetrics.f1])
model.summary()
Model: "Conv1D-word-embed" _________________________________________________________________ Layer (type) Output Shape Param # ================================================================= input_60 (InputLayer) [(None, 1)] 0 _________________________________________________________________ text_vectorization_1 (TextVe (None, 55) 0 _________________________________________________________________ word_embed (Embedding) (None, 55, 128) 8299648 _________________________________________________________________ conv1d_3 (Conv1D) (None, 55, 64) 41024 _________________________________________________________________ global_average_pooling1d_1 ( (None, 64) 0 _________________________________________________________________ dense_56 (Dense) (None, 5) 325 ================================================================= Total params: 8,340,997 Trainable params: 8,340,997 Non-trainable params: 0 _________________________________________________________________
ds = tfdatasets['sent']
(len(ds['train'])*ds['train']._input_dataset._batch_size).numpy()
180064
Since we have close to 180000 data points to train on, even with GPU this would take a long amount of time. To keep the experiments fast enough, we are going to train only on 10% of the samples of these i.e about 18000 samples.
train_steps = int(0.1*len(ds['train']))
val_steps = int(0.1*len(ds['dev']))
model.fit(ds['train'], steps_per_epoch=train_steps,
validation_data=ds['dev'], validation_steps=val_steps,
epochs=NUM_EPOCHS)
MODELS[model_name] = model
Epoch 1/50 562/562 [==============================] - 61s 107ms/step - loss: 0.9116 - accuracy: 0.6391 - f1: 0.5243 - val_loss: 0.6912 - val_accuracy: 0.7377 - val_f1: 0.7072 Epoch 2/50 562/562 [==============================] - 59s 105ms/step - loss: 0.6631 - accuracy: 0.7541 - f1: 0.7302 - val_loss: 0.6335 - val_accuracy: 0.7653 - val_f1: 0.7419 Epoch 3/50 562/562 [==============================] - 59s 105ms/step - loss: 0.6201 - accuracy: 0.7740 - f1: 0.7564 - val_loss: 0.5962 - val_accuracy: 0.7826 - val_f1: 0.7696 Epoch 4/50 562/562 [==============================] - 59s 105ms/step - loss: 0.5890 - accuracy: 0.7889 - f1: 0.7744 - val_loss: 0.5750 - val_accuracy: 0.7926 - val_f1: 0.7821 Epoch 5/50 562/562 [==============================] - 59s 104ms/step - loss: 0.5901 - accuracy: 0.7920 - f1: 0.7790 - val_loss: 0.5581 - val_accuracy: 0.7995 - val_f1: 0.7859 Epoch 6/50 562/562 [==============================] - 59s 105ms/step - loss: 0.5799 - accuracy: 0.7914 - f1: 0.7820 - val_loss: 0.5563 - val_accuracy: 0.8012 - val_f1: 0.7887 Epoch 7/50 562/562 [==============================] - 59s 104ms/step - loss: 0.5584 - accuracy: 0.7999 - f1: 0.7909 - val_loss: 0.5409 - val_accuracy: 0.8059 - val_f1: 0.7954 Epoch 8/50 562/562 [==============================] - 59s 105ms/step - loss: 0.5409 - accuracy: 0.8108 - f1: 0.8022 - val_loss: 0.5271 - val_accuracy: 0.8132 - val_f1: 0.8008 Epoch 9/50 562/562 [==============================] - 58s 104ms/step - loss: 0.5408 - accuracy: 0.8088 - f1: 0.8009 - val_loss: 0.5502 - val_accuracy: 0.7942 - val_f1: 0.7898 Epoch 10/50 562/562 [==============================] - 59s 105ms/step - loss: 0.5439 - accuracy: 0.8058 - f1: 0.7987 - val_loss: 0.5288 - val_accuracy: 0.8068 - val_f1: 0.7984 Epoch 11/50 7/562 [..............................] - ETA: 58s - loss: 0.6573 - accuracy: 0.7750 - f1: 0.7731WARNING:tensorflow:Your input ran out of data; interrupting training. Make sure that your dataset or generator can generate at least `steps_per_epoch * epochs` batches (in this case, 28100 batches). You may need to use the repeat() function when building your dataset.
WARNING:tensorflow:Your input ran out of data; interrupting training. Make sure that your dataset or generator can generate at least `steps_per_epoch * epochs` batches (in this case, 28100 batches). You may need to use the repeat() function when building your dataset.
562/562 [==============================] - 1s 2ms/step - loss: 0.6573 - accuracy: 0.7750 - f1: 0.7731 - val_loss: 0.5404 - val_accuracy: 0.8022 - val_f1: 0.7962
Learning Curve¶
plot_learning_curve(model, extra_metric='accuracy');
Predictions¶
# Make predictions
PREDICTIONS[model_name] = {}
for subset, dset in ds.items():
PREDICTIONS[model_name][subset] = reshape_classification_prediction(model.predict(dset))
Model 2: Universal Sentence Encoder feature extraction¶
- Universal Sentence Encoder takes a whole sentence (we don't need to tokenize it) and turns it into a 512 dimensional embedding
- The paper Neural Networks for Joint Sentence Classification in Medical Paper Abstracts uses pretrained GloVe embedding, and we can also experiment with various different embeddings
The model structure will be as follows:
Input (string) -> Universal Sentence Encoder -> 512 dimensional embedding -> layers -> Output (softmax probabilities)
The method we are following is feature extraction transfer learning
import tensorflow_hub as hub
use_embed_layer = hub.KerasLayer('https://tfhub.dev/google/universal-sentence-encoder/4', trainable=False,
name='universal_sentence_encoder')
use_embed_layer
<tensorflow_hub.keras_layer.KerasLayer at 0x7fe5ab890810>
Let us try with a random sentence
rand_sent = np.random.choice(sentences['train'])
sent_embed = use_embed_layer([rand_sent])
print(f'Text:\n{rand_sent}\n')
print(f'Embedding Shape={sent_embed.shape}:\n', sent_embed)
Text: Conversion from CNI to everolimus to preserve renal function can be considered several years after kidney transplantation and does not compromise immunosuppressive efficacy . Embedding Shape=(1, 512): tf.Tensor( [[ 3.84061108e-03 -4.89690118e-02 -7.75964558e-02 9.30715934e-04 3.63572799e-02 -6.43959045e-02 -8.39816965e-03 -2.59044059e-02 -3.62980999e-02 9.25565977e-03 8.36882889e-02 2.55907252e-02 -6.61989152e-02 1.93596892e-02 7.99917523e-03 -3.31927612e-02 -8.51460546e-02 9.41803120e-03 -8.23286250e-02 5.56989387e-02 -2.89666411e-02 3.02125253e-02 6.40274286e-02 -4.40741070e-02 5.61617129e-02 -3.73874605e-02 2.36412901e-02 7.16379210e-02 -6.00813255e-02 -1.51469058e-03 -5.38987331e-02 6.69857487e-02 3.17833126e-02 6.19302830e-03 -8.00436828e-03 -6.72090575e-02 -5.42498864e-02 4.39067222e-02 -1.16901789e-02 7.24743456e-02 6.17060810e-02 -7.62524009e-02 -2.51462348e-02 -6.01319075e-02 -2.92660333e-02 5.45057841e-02 -6.60429662e-03 -4.40517031e-02 4.35051061e-02 2.22238172e-02 -4.51276451e-02 -5.38100116e-02 -4.51281071e-02 3.17592882e-02 -4.00199704e-02 4.44068313e-02 3.35098729e-02 -3.37215960e-02 7.21965581e-02 -3.82919461e-02 -3.20348665e-02 6.94982335e-02 5.43962512e-03 -5.67801632e-02 1.02294832e-02 -1.13122826e-02 -2.38631349e-02 2.81251781e-02 1.30594028e-02 4.66825347e-03 -3.21429893e-02 -1.64098050e-02 7.51353800e-02 8.09700345e-04 1.35874469e-02 5.95752634e-02 2.54117362e-02 6.22052960e-02 1.65239777e-02 4.78524715e-02 -6.34682029e-02 5.25586419e-02 3.85677740e-02 5.03873006e-02 -6.59354627e-02 -8.50478373e-03 2.31860932e-02 2.93434951e-02 6.78832317e-03 2.58442853e-02 1.96010191e-02 -4.57845889e-02 -5.28330691e-02 1.55332936e-02 -6.81953877e-02 -4.54576053e-02 -5.87696433e-02 2.67114434e-02 6.35554418e-02 -7.12315366e-02 -7.50472248e-02 -4.61141802e-02 -5.91964200e-02 3.38927545e-02 -4.06114161e-02 -6.97994325e-03 2.63259951e-02 5.44207245e-02 -3.82102951e-02 4.79041114e-02 -7.08463863e-02 -2.76543610e-02 7.21288437e-04 -1.58719886e-02 -3.78736667e-02 -1.98269752e-03 -4.80769947e-02 -4.50404398e-02 5.75538212e-03 2.51387488e-02 5.11586964e-02 -6.28072619e-02 1.28243444e-02 4.54736985e-02 2.98069157e-02 -4.53450866e-02 -3.56083475e-02 1.58106834e-02 6.33833483e-02 -7.32476860e-02 3.97247039e-02 8.42631161e-02 8.67618527e-03 -2.36487351e-02 -1.33573301e-02 -2.79972702e-02 5.16036302e-02 -5.73783927e-02 -3.02799661e-02 -7.13946391e-03 -6.26772270e-02 3.43643427e-02 1.95036978e-02 2.97193266e-02 -4.96654324e-02 -2.57291704e-05 -1.66486427e-02 -7.11807311e-02 -2.42565665e-02 -8.21643323e-03 -5.89324348e-02 -1.16085829e-02 -6.78637028e-02 6.10072799e-02 7.13245794e-02 1.24281133e-02 2.46704072e-02 4.75547090e-02 -2.53095366e-02 -2.86618453e-02 2.79791821e-02 -6.06752140e-03 -3.29410210e-02 4.91629653e-02 -2.99548469e-02 -6.28587678e-02 -4.60708253e-02 1.59241986e-02 1.88582335e-02 -4.82132658e-02 -2.10544225e-02 -8.17153826e-02 2.66442634e-02 -8.22690278e-02 1.67708956e-02 -6.07919991e-02 3.79176741e-03 -6.42280728e-02 -2.42040362e-02 -1.87737923e-02 -7.57839158e-02 1.19734490e-02 3.23401168e-02 2.61170063e-02 4.97125613e-04 2.44905353e-02 -4.12557414e-03 2.99274251e-02 3.17453556e-02 -1.17841763e-02 -3.86062376e-02 -6.61664410e-03 4.90230955e-02 2.01377552e-02 5.25683984e-02 -8.05467833e-03 4.65246402e-02 -1.45302620e-02 2.66212281e-02 -2.81894263e-02 -4.61146235e-03 4.92718397e-03 -8.19239318e-02 5.93146570e-02 2.59505715e-02 -5.18163964e-02 3.02839708e-02 -5.89410141e-02 7.00150756e-03 7.11098313e-02 -6.15194552e-02 3.79615575e-02 4.61568758e-02 1.59254186e-02 -2.49365345e-02 -4.31363843e-02 -4.55422513e-02 -1.74011253e-02 2.73718368e-02 -3.04597616e-02 5.52191995e-02 -5.23021594e-02 1.28098167e-02 8.79738759e-03 2.99134739e-02 7.25900242e-03 -3.04852221e-02 1.41027719e-02 6.47329837e-02 2.46176403e-02 -7.00403079e-02 4.44220975e-02 -7.35673308e-02 -7.77102262e-02 8.74995440e-03 1.69080570e-02 2.11022887e-03 -7.34883547e-02 -4.28707637e-02 4.03814688e-02 -1.69983646e-03 5.09600947e-03 -5.64582869e-02 -6.30637407e-02 8.88175983e-03 -2.22277567e-02 -6.04570052e-03 3.84855829e-02 2.83059143e-02 -7.44221061e-02 -4.86048982e-02 -6.63966686e-02 3.33658382e-02 -2.28838734e-02 -3.48658450e-02 2.01750081e-02 -5.39638065e-02 5.60753271e-02 -6.36855587e-02 1.09851416e-02 8.31264909e-03 5.54529354e-02 -7.59856999e-02 -2.32037213e-02 -8.61131586e-03 -6.74034134e-02 4.77494709e-02 -4.72037382e-02 -1.10528972e-02 -4.78951111e-02 -5.76296337e-02 -4.22491282e-02 6.78630024e-02 -5.10197543e-02 5.33100441e-02 -4.75425124e-02 6.55710250e-02 -3.32144871e-02 5.28863072e-02 -4.55092303e-02 4.21004668e-02 9.22758039e-03 -4.02246937e-02 1.06170764e-02 -2.66237631e-02 -8.28400720e-03 -7.30094090e-02 -4.66944128e-02 -4.11690436e-02 3.12028769e-02 4.65516783e-02 3.17948982e-02 -1.56653263e-02 6.45347163e-02 5.68211675e-02 -1.93077624e-02 -6.46382496e-02 -9.75340512e-03 -4.78193983e-02 7.24242628e-02 4.14608754e-02 7.72337243e-03 -2.09880266e-02 -7.17649385e-02 4.57113162e-02 -1.61594693e-02 -5.89481601e-03 -5.30919172e-02 -6.13906607e-02 1.18486406e-02 -4.22424264e-02 -4.48050499e-02 -8.25610906e-02 3.64829265e-02 -3.25715356e-02 5.75227961e-02 -4.54122983e-02 -5.44589125e-02 3.54822017e-02 4.80801277e-02 5.59169725e-02 -5.42256869e-02 -7.23249093e-02 -5.23786992e-02 -5.69901057e-03 7.57505149e-02 -7.76385739e-02 1.74277984e-02 -3.11719812e-02 -7.07057714e-02 -1.23999361e-02 -3.50238532e-02 3.76466066e-02 7.28575140e-02 6.20676065e-03 -1.67576410e-02 -4.27524708e-02 2.61412207e-02 2.00443789e-02 5.87267019e-02 3.13566625e-02 5.22531271e-02 -1.74065698e-02 4.59137708e-02 -6.38068467e-02 -1.47033231e-02 2.35591512e-02 -2.57369783e-02 -6.85043260e-02 4.61189672e-02 2.61171884e-03 -1.82350315e-02 -5.04288040e-02 -3.61958705e-02 -1.84727926e-02 2.38842145e-02 -6.80451393e-02 -6.26738816e-02 5.83330654e-02 -1.98964346e-02 -5.64292446e-02 -4.24523875e-02 -3.35064195e-02 4.57418198e-03 1.56283919e-02 -4.69899699e-02 6.29458353e-02 3.14078107e-02 -4.89341505e-02 7.31584728e-02 -1.12847416e-02 7.89217651e-02 -3.32480110e-02 -2.79928558e-02 -1.26591632e-02 -6.01822101e-02 -8.43352638e-03 -4.88049164e-02 1.87628735e-02 2.21437234e-02 -5.30426055e-02 2.63753142e-02 7.49413436e-03 -6.30301908e-02 6.57100454e-02 4.15280014e-02 -4.05803621e-02 5.69360293e-02 7.46220648e-02 -2.43262276e-02 -1.26542675e-03 -3.40649113e-02 3.04252356e-02 -8.25234801e-02 5.60104884e-02 3.32329236e-02 6.46687821e-02 4.60800482e-03 2.74764318e-02 3.22911553e-02 7.38011450e-02 -1.30111668e-02 -1.90809648e-02 -1.14450185e-02 -1.42365173e-02 5.11149764e-02 5.94341122e-02 2.36219596e-02 -4.71569113e-02 -5.40824840e-03 2.88420208e-02 4.63770516e-03 6.24290220e-02 -4.76743355e-02 -6.26409799e-02 9.06704087e-03 5.69748916e-02 -3.44932713e-02 6.86231405e-02 -1.45627726e-02 -1.03601639e-03 -3.76393609e-02 5.75770042e-04 -3.15601118e-02 -6.18663020e-02 -2.91718319e-02 3.87815535e-02 2.70903762e-02 5.94319329e-02 1.78584573e-03 -6.21453896e-02 -5.35957776e-02 -2.19953693e-02 5.35643585e-02 -5.67859560e-02 4.21736203e-02 2.27596890e-02 1.94445252e-02 -3.32135893e-02 4.95323613e-02 -3.99950221e-02 7.77157769e-03 -1.87146887e-02 3.79665494e-02 4.52085473e-02 -5.56367487e-02 5.55064939e-02 8.24468303e-03 -8.21523890e-02 -9.47243534e-03 5.51270582e-02 -3.30185294e-02 -2.79286411e-02 2.79778149e-02 -5.73564172e-02 -3.60449106e-02 -7.60263577e-02 3.37580293e-02 -5.04767038e-02 4.63643447e-02 4.80567180e-02 4.80425954e-02 1.10993199e-02 -5.93237616e-02 1.47754485e-02 -5.15358010e-03 -8.33996758e-02 -3.69450264e-02 -5.97986653e-02 -7.36954436e-03 -4.65769880e-02 5.18256351e-02 -2.67247157e-03 -5.01645952e-02 3.54410731e-03 3.76989990e-02 4.00882587e-02 2.62877773e-02 3.35277990e-02 4.26940471e-02 -7.89648741e-02 7.80914351e-02 2.74627786e-02 -4.86164317e-02 -6.66435957e-02 3.73356007e-02 -5.00042774e-02 -5.86151611e-03 -4.59308876e-03 -1.14124641e-02 6.22379184e-02 1.25738513e-03 -7.85934106e-02 -1.09839356e-02 4.70221303e-02 -1.14967525e-02 7.42812753e-02 -4.85409535e-02 6.19559214e-02 -5.36380894e-03 -1.13996258e-02 1.31332520e-02 -6.95186779e-02 -6.31158501e-02 5.79894483e-02 -8.31967443e-02 -2.13006599e-04 4.83379699e-02 -4.76069897e-02 1.15615409e-02 7.68028572e-03 -6.96112663e-02]], shape=(1, 512), dtype=float32)
🔑 Note:
- USE returns a 512 dimensional vector for the entire sentence.
- The embedding layer which we used earlier would return a d-dimensional embedding for each separate token, and hence if there are n tokens in the sentence, the output shape would be (num_tokens, embed_dim)
Creating the model¶
model_name = 'USE-feature-extraction'
inputs = layers.Input(shape=[], dtype=tf.string)
embedding = use_embed_layer(inputs)
x = layers.Dense(128, activation='relu')(embedding)
outputs = layers.Dense(N_CLASSES, activation='softmax')(x)
model = tf.keras.models.Model(inputs, outputs, name=model_name)
model.compile(loss='categorical_crossentropy', optimizer=tf.keras.optimizers.Adam(),
metrics=['accuracy', KerasMetrics.f1])
model.summary()
Model: "USE-feature-extraction" _________________________________________________________________ Layer (type) Output Shape Param # ================================================================= input_61 (InputLayer) [(None,)] 0 _________________________________________________________________ universal_sentence_encoder ( (None, 512) 256797824 _________________________________________________________________ dense_57 (Dense) (None, 128) 65664 _________________________________________________________________ dense_58 (Dense) (None, 5) 645 ================================================================= Total params: 256,864,133 Trainable params: 66,309 Non-trainable params: 256,797,824 _________________________________________________________________
ds = tfdatasets['sent']
train_steps = int(0.1*len(ds['train']))
val_steps = int(0.1*len(ds['dev']))
model.fit(ds['train'], steps_per_epoch=train_steps,
validation_data=ds['dev'], validation_steps=val_steps,
epochs=NUM_EPOCHS)
MODELS[model_name] = model
Epoch 1/50 562/562 [==============================] - 13s 18ms/step - loss: 0.9182 - accuracy: 0.6480 - f1: 0.5613 - val_loss: 0.7966 - val_accuracy: 0.6898 - val_f1: 0.6695 Epoch 2/50 562/562 [==============================] - 9s 16ms/step - loss: 0.7674 - accuracy: 0.7028 - f1: 0.6830 - val_loss: 0.7545 - val_accuracy: 0.7045 - val_f1: 0.6925 Epoch 3/50 562/562 [==============================] - 10s 17ms/step - loss: 0.7496 - accuracy: 0.7130 - f1: 0.6987 - val_loss: 0.7361 - val_accuracy: 0.7158 - val_f1: 0.6969 Epoch 4/50 562/562 [==============================] - 9s 17ms/step - loss: 0.7151 - accuracy: 0.7259 - f1: 0.7107 - val_loss: 0.7077 - val_accuracy: 0.7294 - val_f1: 0.7180 Epoch 5/50 562/562 [==============================] - 9s 16ms/step - loss: 0.7233 - accuracy: 0.7224 - f1: 0.7087 - val_loss: 0.6871 - val_accuracy: 0.7350 - val_f1: 0.7212 Epoch 6/50 562/562 [==============================] - 10s 17ms/step - loss: 0.7142 - accuracy: 0.7268 - f1: 0.7124 - val_loss: 0.6807 - val_accuracy: 0.7340 - val_f1: 0.7187 Epoch 7/50 562/562 [==============================] - 9s 17ms/step - loss: 0.6847 - accuracy: 0.7392 - f1: 0.7245 - val_loss: 0.6643 - val_accuracy: 0.7483 - val_f1: 0.7363 Epoch 8/50 562/562 [==============================] - 9s 17ms/step - loss: 0.6729 - accuracy: 0.7431 - f1: 0.7341 - val_loss: 0.6530 - val_accuracy: 0.7487 - val_f1: 0.7388 Epoch 9/50 562/562 [==============================] - 9s 17ms/step - loss: 0.6713 - accuracy: 0.7432 - f1: 0.7329 - val_loss: 0.6562 - val_accuracy: 0.7497 - val_f1: 0.7364 Epoch 10/50 562/562 [==============================] - 9s 17ms/step - loss: 0.6666 - accuracy: 0.7470 - f1: 0.7346 - val_loss: 0.6498 - val_accuracy: 0.7580 - val_f1: 0.7418 Epoch 11/50 5/562 [..............................] - ETA: 8s - loss: 0.6785 - accuracy: 0.7937 - f1: 0.7672WARNING:tensorflow:Your input ran out of data; interrupting training. Make sure that your dataset or generator can generate at least `steps_per_epoch * epochs` batches (in this case, 28100 batches). You may need to use the repeat() function when building your dataset.
WARNING:tensorflow:Your input ran out of data; interrupting training. Make sure that your dataset or generator can generate at least `steps_per_epoch * epochs` batches (in this case, 28100 batches). You may need to use the repeat() function when building your dataset.
562/562 [==============================] - 1s 2ms/step - loss: 0.6951 - accuracy: 0.7700 - f1: 0.7210 - val_loss: 0.6529 - val_accuracy: 0.7550 - val_f1: 0.7419
Learning curve¶
plot_learning_curve(model, extra_metric='accuracy');
Predictions¶
# Make predictions
PREDICTIONS[model_name] = {}
for subset, dset in ds.items():
PREDICTIONS[model_name][subset] = reshape_classification_prediction(model.predict(dset))
Model 3: Conv1D with character embeddings¶
Creating a character level tokenizer¶
- The paper Neural Networks for Joint Sentence Classification in Medical Paper Abstracts uses a combination of word-level and character embeddings (hybrid-concatenated)
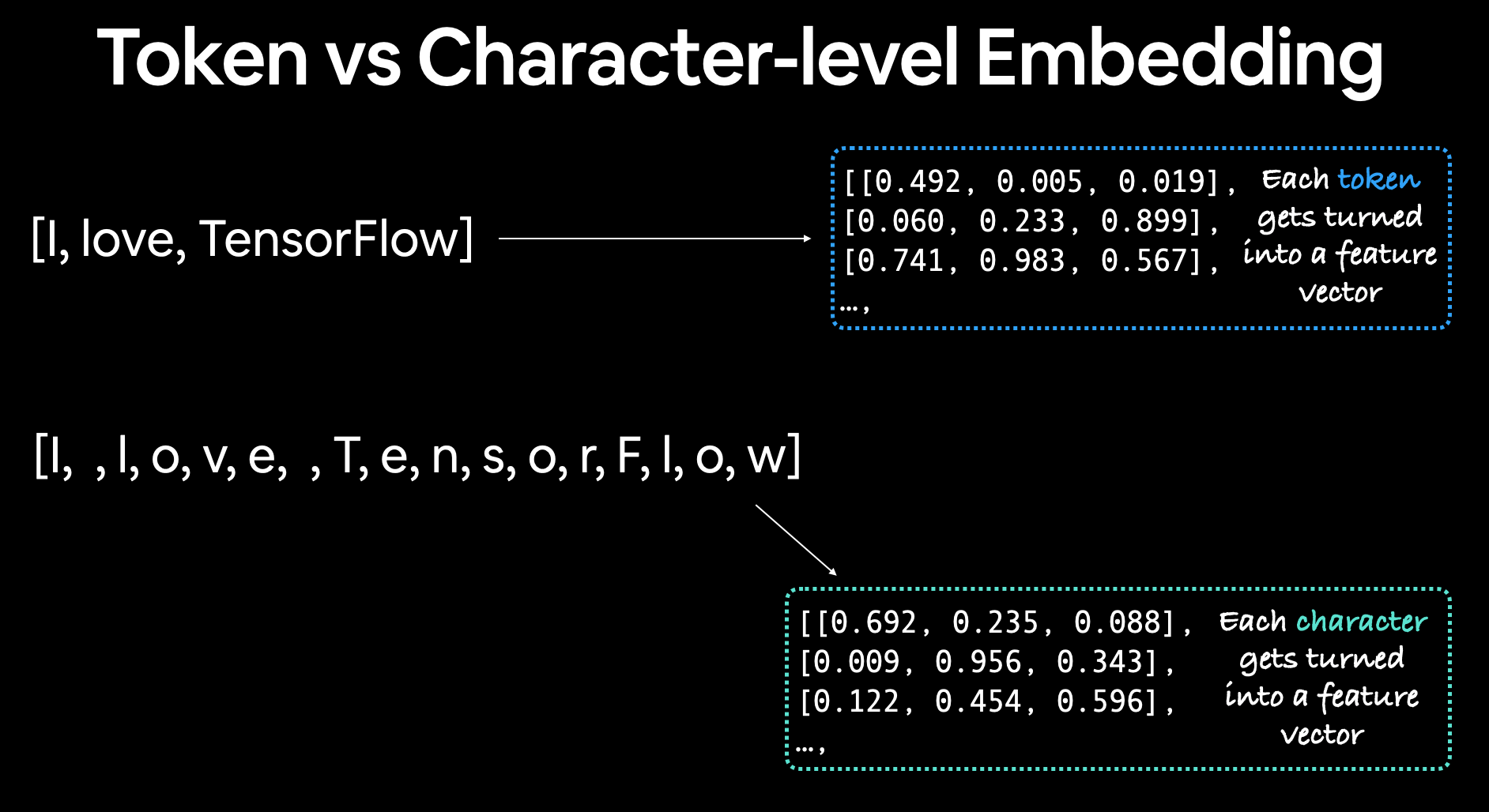
Correction for above image:
- In word-level tokenization, each word is a separate token, and in character-level tokenization each character is a separate token. Both are ultimately called tokens.
- Each token gets its own feature vector aka embedding.
Note:
- The vocabulary length will be much much higher in word-level tokenization than character-level tokenization
- But the sequence length in character-level tokenization will be much much higher than in word-level tokenization (This can cause problems with sequence models such as LSTM, due to extra long sequences and model not being able to remember long-range dependencies)
def split_chars(text, return_string=False):
chars = list(text)
if return_string:
return ' '.join(chars)
return chars
characters = {}
for subset in ['train', 'dev', 'test']:
characters[subset] = [split_chars(sent, return_string=True) for sent in sentences[subset]]
rand_sent = np.random.choice(sentences['train'])
sent_chars = split_chars(rand_sent, return_string=True)
print(f'Text:\n{rand_sent}\n')
print(f'Characters:\n{sent_chars}')
Text: The @ pair of bilateral acupoints were fixed with self-adhesive electrodes and connected with Han 's acupoint and nerve stimulator ( HANS , LH@H ) , the frequency was @ Hz / @ Hz , the intensity was @ - @ mA and the form was densedisperse wave within the patients ' tolarance . Characters: T h e @ p a i r o f b i l a t e r a l a c u p o i n t s w e r e f i x e d w i t h s e l f - a d h e s i v e e l e c t r o d e s a n d c o n n e c t e d w i t h H a n ' s a c u p o i n t a n d n e r v e s t i m u l a t o r ( H A N S , L H @ H ) , t h e f r e q u e n c y w a s @ H z / @ H z , t h e i n t e n s i t y w a s @ - @ m A a n d t h e f o r m w a s d e n s e d i s p e r s e w a v e w i t h i n t h e p a t i e n t s ' t o l a r a n c e .
Let us look at some character-level token statistics
sent_char_lens = [len(sent) for sent in sentences['train']]
np.mean(sent_char_lens)
149.3662574983337
plt.hist(sent_char_lens, bins=10);
Most of the sentences have character lengths between 250 and 300ish. Let us see the 95 percentile, so that we can limit the maximum sequence length
max_seq_length = int(np.percentile(sent_char_lens, 95))
max_seq_length
290
Now let us limit the character level vocab to 26 ascii (lowercase) characters and 2 extra tokens (space, OOV)
max_vocab_length = 26 + 2
char_vectorizer = TextVectorization(max_tokens=max_vocab_length,
standardize='lower_and_strip_punctuation',
split='whitespace',
output_sequence_length=max_seq_length,
name='char_vectorizer')
# Adapt to the training set
char_vectorizer.adapt(characters['train'])
vocab = char_vectorizer.get_vocabulary()
print(f'Number of tokens in the vocab: {len(vocab)}')
print(f'5 most common tokens:', vocab[:5])
print(f'5 least common tokens:', vocab[-5:])
Number of tokens in the vocab: 28 5 most common tokens: ['', '[UNK]', 'e', 't', 'i'] 5 least common tokens: ['k', 'x', 'z', 'q', 'j']
Let's test it on a random sentence
subset = 'train'
idx = np.random.randint(0, len(sentences[subset]))
rand_sent = sentences[subset][idx]
rand_sent_chars = characters[subset][idx]
char_vec = char_vectorizer([rand_sent_chars])
print(f'Sentence:\n{rand_sent}\n')
print(f'Characters:\n{rand_sent_chars}\n')
print(f'Vectorized:\n{char_vec}\n')
Sentence: New drug resistance to AA was not seen . Characters: N e w d r u g r e s i s t a n c e t o A A w a s n o t s e e n . Vectorized: [[ 6 2 20 10 8 16 18 8 2 9 4 9 3 5 6 11 2 3 7 5 5 20 5 9 6 7 3 9 2 2 6 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0]]
Create a character level embedding¶
char_embed = layers.Embedding(input_dim=len(vocab),
output_dim=25, # Learning a 25-dimensional embedding for each token in vocab
mask_zero=True,
name='char_embed')
subset = 'train'
idx = np.random.randint(0, len(sentences[subset]))
rand_sent = sentences[subset][idx]
rand_sent_chars = characters[subset][idx]
char_vec = char_vectorizer([rand_sent_chars])
char_embedding = char_embed(char_vec)
print(f'Sentence:\n{rand_sent}\n')
print(f'Characters:\n{rand_sent_chars}\n')
print(f'Vectorized Shape={char_vec.shape}:\n{char_vec}\n')
print(f'Embedding Shape={char_embedding.shape}:\n{char_embedding}\n')
Sentence:
AbbVie , Gutsy Group , Gandel Philanthropy , Angior Foundation , Crohn 's Colitis Australia , and the National Health and Medical Research Council .
Characters:
A b b V i e , G u t s y G r o u p , G a n d e l P h i l a n t h r o p y , A n g i o r F o u n d a t i o n , C r o h n ' s C o l i t i s A u s t r a l i a , a n d t h e N a t i o n a l H e a l t h a n d M e d i c a l R e s e a r c h C o u n c i l .
Vectorized Shape=(1, 290):
[[ 5 22 22 21 4 2 18 16 3 9 19 18 8 7 16 14 18 5 6 10 2 12 14 13
4 12 5 6 3 13 8 7 14 19 5 6 18 4 7 8 17 7 16 6 10 5 3 4
7 6 11 8 7 13 6 9 11 7 12 4 3 4 9 5 16 9 3 8 5 12 4 5
5 6 10 3 13 2 6 5 3 4 7 6 5 12 13 2 5 12 3 13 5 6 10 15
2 10 4 11 5 12 8 2 9 2 5 8 11 13 11 7 16 6 11 4 12 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0]]
Embedding Shape=(1, 290, 25):
[[[ 7.9680234e-05 -4.5517575e-02 1.3455022e-02 ... -4.5774888e-02
-2.4014855e-02 3.8422409e-02]
[ 4.1982904e-03 4.7290329e-02 -6.3347928e-03 ... 1.3557676e-02
-3.2506216e-02 -1.0842133e-02]
[ 4.1982904e-03 4.7290329e-02 -6.3347928e-03 ... 1.3557676e-02
-3.2506216e-02 -1.0842133e-02]
...
[ 3.7082221e-02 4.8321847e-02 4.0627506e-02 ... -4.4790257e-02
5.7347864e-04 2.3226772e-02]
[ 3.7082221e-02 4.8321847e-02 4.0627506e-02 ... -4.4790257e-02
5.7347864e-04 2.3226772e-02]
[ 3.7082221e-02 4.8321847e-02 4.0627506e-02 ... -4.4790257e-02
5.7347864e-04 2.3226772e-02]]]
Making the TensorSliceDataset and PrefetchDataset¶
for subset in ['train', 'dev', 'test']:
tfdatasets['char'][subset] = tf.data.Dataset.from_tensor_slices((characters[subset], data_labels['one_hot'][subset]))
tfdatasets['char']['train']
<TensorSliceDataset shapes: ((), (5,)), types: (tf.string, tf.float64)>
Now, batch and prefetch
for subset in ['train', 'dev', 'test']:
tfdatasets['char'][subset] = tfdatasets['char'][subset].batch(32).prefetch(tf.data.AUTOTUNE)
tfdatasets['char']['train']
<PrefetchDataset shapes: ((None,), (None, 5)), types: (tf.string, tf.float64)>
This is how our model structure will look like:
Input (string) -> Tokenize (character-level) -> Embedding (25-dimensional) -> layers -> Output (softmax probability)model_name = 'Conv1D-char-embed'
inputs = layers.Input(shape=(1,), dtype=tf.string)
char_vectors = char_vectorizer(inputs)
char_embeddings = char_embed(char_vectors)
x = layers.Conv1D(64, kernel_size=5, padding='same', activation='relu')(char_embeddings)
x = layers.GlobalMaxPool1D()(x)
outputs = layers.Dense(N_CLASSES, activation='softmax')(x)
model = tf.keras.models.Model(inputs, outputs, name=model_name)
model.compile(loss='categorical_crossentropy', optimizer=tf.keras.optimizers.Adam(),
metrics=['accuracy', KerasMetrics.f1])
model.summary()
Model: "Conv1D-char-embed" _________________________________________________________________ Layer (type) Output Shape Param # ================================================================= input_62 (InputLayer) [(None, 1)] 0 _________________________________________________________________ char_vectorizer (TextVectori (None, 290) 0 _________________________________________________________________ char_embed (Embedding) (None, 290, 25) 700 _________________________________________________________________ conv1d_4 (Conv1D) (None, 290, 64) 8064 _________________________________________________________________ global_max_pooling1d_2 (Glob (None, 64) 0 _________________________________________________________________ dense_59 (Dense) (None, 5) 325 ================================================================= Total params: 9,089 Trainable params: 9,089 Non-trainable params: 0 _________________________________________________________________
ds = tfdatasets['char']
train_steps = int(0.1*len(ds['train']))
val_steps = int(0.1*len(ds['dev']))
model.fit(ds['train'], steps_per_epoch=train_steps,
validation_data=ds['dev'], validation_steps=val_steps,
epochs=NUM_EPOCHS)
MODELS[model_name] = model
Epoch 1/50 562/562 [==============================] - 8s 9ms/step - loss: 1.2602 - accuracy: 0.4922 - f1: 0.2587 - val_loss: 1.0439 - val_accuracy: 0.5841 - val_f1: 0.4693 Epoch 2/50 562/562 [==============================] - 5s 9ms/step - loss: 1.0042 - accuracy: 0.5963 - f1: 0.5310 - val_loss: 0.9400 - val_accuracy: 0.6287 - val_f1: 0.5774 Epoch 3/50 562/562 [==============================] - 5s 8ms/step - loss: 0.9284 - accuracy: 0.6371 - f1: 0.5876 - val_loss: 0.8661 - val_accuracy: 0.6705 - val_f1: 0.6266 Epoch 4/50 562/562 [==============================] - 5s 8ms/step - loss: 0.8751 - accuracy: 0.6595 - f1: 0.6208 - val_loss: 0.8408 - val_accuracy: 0.6805 - val_f1: 0.6384 Epoch 5/50 562/562 [==============================] - 5s 8ms/step - loss: 0.8583 - accuracy: 0.6658 - f1: 0.6361 - val_loss: 0.8217 - val_accuracy: 0.6898 - val_f1: 0.6567 Epoch 6/50 562/562 [==============================] - 5s 9ms/step - loss: 0.8390 - accuracy: 0.6755 - f1: 0.6443 - val_loss: 0.7947 - val_accuracy: 0.6961 - val_f1: 0.6609 Epoch 7/50 562/562 [==============================] - 5s 9ms/step - loss: 0.8256 - accuracy: 0.6783 - f1: 0.6519 - val_loss: 0.7878 - val_accuracy: 0.6995 - val_f1: 0.6674 Epoch 8/50 562/562 [==============================] - 5s 8ms/step - loss: 0.7941 - accuracy: 0.6927 - f1: 0.6720 - val_loss: 0.7892 - val_accuracy: 0.6958 - val_f1: 0.6782 Epoch 9/50 562/562 [==============================] - 5s 8ms/step - loss: 0.7942 - accuracy: 0.6952 - f1: 0.6719 - val_loss: 0.7854 - val_accuracy: 0.6908 - val_f1: 0.6792 Epoch 10/50 562/562 [==============================] - 5s 8ms/step - loss: 0.7888 - accuracy: 0.6960 - f1: 0.6745 - val_loss: 0.7480 - val_accuracy: 0.7151 - val_f1: 0.6897 Epoch 11/50 7/562 [..............................] - ETA: 4s - loss: 0.8328 - accuracy: 0.6700 - f1: 0.6491WARNING:tensorflow:Your input ran out of data; interrupting training. Make sure that your dataset or generator can generate at least `steps_per_epoch * epochs` batches (in this case, 28100 batches). You may need to use the repeat() function when building your dataset.
WARNING:tensorflow:Your input ran out of data; interrupting training. Make sure that your dataset or generator can generate at least `steps_per_epoch * epochs` batches (in this case, 28100 batches). You may need to use the repeat() function when building your dataset.
562/562 [==============================] - 1s 891us/step - loss: 0.8328 - accuracy: 0.6700 - f1: 0.6491 - val_loss: 0.7647 - val_accuracy: 0.6975 - val_f1: 0.6798
Learning Curve¶
plot_learning_curve(model, extra_metric='accuracy');
Predictions¶
# Make predictions
PREDICTIONS[model_name] = {}
for subset, dset in ds.items():
PREDICTIONS[model_name][subset] = reshape_classification_prediction(model.predict(dset))
Model 4: Combining pretrained word embeddings + character embeddings (hybrid embedding layer)¶
We are moving closer to the architechture presented in Figure 1 in the paper Neural Networks for Joint Sentence Classification in Medical Paper Abstracts (See below in Model 5)
We are going to make a hybrid embedding by concatenating the pretrained embedding (from USE), with a character level embedding (which will be trainable) - by stacking/concatenating.
To make this archiechture:
- Make the use_embedding_layer as in model 2
- Make the character embedding layer as in model 3 -> Pass it through some non-linear layers to learn a more concatenable representation
- Use
layers.concatenateto concatenate the outputs of 1 and 2 - Now add some hidden layers
- Finally add the output layer
model_name = 'USE-char-hybrid-embed'
# USE embeddings model
use_model_inputs = layers.Input(shape=[], dtype=tf.string)
sent_embed = use_embed_layer(use_model_inputs)
use_model_outputs = layers.Dense(128, activation='relu')(sent_embed)
use_model = tf.keras.models.Model(use_model_inputs, use_model_outputs)
# Character embeddings model
char_model_inputs = layers.Input(shape=(1,), dtype=tf.string)
char_vectorized = char_vectorizer(char_model_inputs)
char_embedding = char_embed(char_vectorized)
char_bi_lstm = layers.Bidirectional(layers.LSTM(25))(char_embedding)
char_model = tf.keras.models.Model(char_model_inputs, char_bi_lstm)
# Concatenation of embeddings
embed_concat = layers.Concatenate(name='embed_concatenate_layer')([use_model.output, char_model.output])
# More Hidden layers
initial_dropout = layers.Dropout(0.5)(embed_concat)
dense = layers.Dense(200, activation='relu')(initial_dropout)
final_dropout = layers.Dropout(0.5)(dense)
outputs = layers.Dense(N_CLASSES, activation='softmax')(final_dropout)
# Put together the inputs and the outputs
model = tf.keras.models.Model(inputs=[use_model.inputs, char_model.inputs],
outputs=outputs, name=model_name)
# Compile
model.compile(loss='categorical_crossentropy', optimizer=tf.keras.optimizers.Adam(),
metrics=['accuracy', KerasMetrics.f1])
# Summary
model.summary()
Model: "USE-char-hybrid-embed"
__________________________________________________________________________________________________
Layer (type) Output Shape Param # Connected to
==================================================================================================
input_64 (InputLayer) [(None, 1)] 0
__________________________________________________________________________________________________
input_63 (InputLayer) [(None,)] 0
__________________________________________________________________________________________________
char_vectorizer (TextVectorizat (None, 290) 0 input_64[0][0]
__________________________________________________________________________________________________
universal_sentence_encoder (Ker (None, 512) 256797824 input_63[0][0]
__________________________________________________________________________________________________
char_embed (Embedding) (None, 290, 25) 700 char_vectorizer[1][0]
__________________________________________________________________________________________________
dense_60 (Dense) (None, 128) 65664 universal_sentence_encoder[1][0]
__________________________________________________________________________________________________
bidirectional_12 (Bidirectional (None, 50) 10200 char_embed[1][0]
__________________________________________________________________________________________________
embed_concatenate_layer (Concat (None, 178) 0 dense_60[0][0]
bidirectional_12[0][0]
__________________________________________________________________________________________________
dropout_10 (Dropout) (None, 178) 0 embed_concatenate_layer[0][0]
__________________________________________________________________________________________________
dense_61 (Dense) (None, 200) 35800 dropout_10[0][0]
__________________________________________________________________________________________________
dropout_11 (Dropout) (None, 200) 0 dense_61[0][0]
__________________________________________________________________________________________________
dense_62 (Dense) (None, 5) 1005 dropout_11[0][0]
==================================================================================================
Total params: 256,911,193
Trainable params: 113,369
Non-trainable params: 256,797,824
__________________________________________________________________________________________________
Let's see how the model architecture looks like
tf.keras.utils.plot_model(model, to_file='/tmp/model.png')
Creating a specific tf.data.Dataset for this model¶
- Again, we do this to speed up the model input ingestion process as
tf.data.DatasetAPI provides efficient mapping, prefetching, and batching of data.
sent_char_hybrid_dataset = {}
for subset in ['train', 'dev', 'test']:
# Make the tfdata
inputs_tfdata = tf.data.Dataset.from_tensor_slices((sentences[subset], characters[subset]))
labels_tfdata = tf.data.Dataset.from_tensor_slices(data_labels['one_hot'][subset])
tfdata = tf.data.Dataset.zip((inputs_tfdata, labels_tfdata))
# prefetch and batch
tfdata = tfdata.batch(32).prefetch(buffer_size=tf.data.AUTOTUNE)
sent_char_hybrid_dataset[subset] = tfdata
sent_char_hybrid_dataset['train']
<PrefetchDataset shapes: (((None,), (None,)), (None, 5)), types: ((tf.string, tf.string), tf.float64)>
Now let us the fit the model¶
ds = sent_char_hybrid_dataset
train_steps = int(0.1*len(ds['train']))
val_steps = int(0.1*len(ds['dev']))
model.fit(ds['train'], steps_per_epoch=train_steps, validation_data=ds['dev'],
epochs=NUM_EPOCHS)
MODELS[model_name] = model
Epoch 1/50 562/562 [==============================] - 70s 107ms/step - loss: 0.9595 - accuracy: 0.6187 - f1: 0.5455 - val_loss: 0.7728 - val_accuracy: 0.6991 - val_f1: 0.6759 Epoch 2/50 562/562 [==============================] - 57s 101ms/step - loss: 0.7816 - accuracy: 0.6965 - f1: 0.6743 - val_loss: 0.7074 - val_accuracy: 0.7324 - val_f1: 0.7137 Epoch 3/50 562/562 [==============================] - 53s 95ms/step - loss: 0.7583 - accuracy: 0.7125 - f1: 0.6947 - val_loss: 0.6841 - val_accuracy: 0.7378 - val_f1: 0.7214 Epoch 4/50 562/562 [==============================] - 60s 106ms/step - loss: 0.7316 - accuracy: 0.7217 - f1: 0.7036 - val_loss: 0.6662 - val_accuracy: 0.7478 - val_f1: 0.7334 Epoch 5/50 562/562 [==============================] - 56s 99ms/step - loss: 0.7360 - accuracy: 0.7203 - f1: 0.7018 - val_loss: 0.6547 - val_accuracy: 0.7491 - val_f1: 0.7389 Epoch 6/50 562/562 [==============================] - 55s 98ms/step - loss: 0.7330 - accuracy: 0.7194 - f1: 0.7018 - val_loss: 0.6523 - val_accuracy: 0.7524 - val_f1: 0.7364 Epoch 7/50 562/562 [==============================] - 60s 106ms/step - loss: 0.7065 - accuracy: 0.7326 - f1: 0.7192 - val_loss: 0.6378 - val_accuracy: 0.7578 - val_f1: 0.7475 Epoch 8/50 562/562 [==============================] - 56s 100ms/step - loss: 0.6916 - accuracy: 0.7379 - f1: 0.7265 - val_loss: 0.6258 - val_accuracy: 0.7616 - val_f1: 0.7529 Epoch 9/50 562/562 [==============================] - 56s 100ms/step - loss: 0.6965 - accuracy: 0.7352 - f1: 0.7233 - val_loss: 0.6328 - val_accuracy: 0.7576 - val_f1: 0.7484 Epoch 10/50 562/562 [==============================] - 57s 102ms/step - loss: 0.6916 - accuracy: 0.7366 - f1: 0.7232 - val_loss: 0.6184 - val_accuracy: 0.7645 - val_f1: 0.7553 Epoch 11/50 7/562 [..............................] - ETA: 26s - loss: 0.7348 - accuracy: 0.7100 - f1: 0.6739WARNING:tensorflow:Your input ran out of data; interrupting training. Make sure that your dataset or generator can generate at least `steps_per_epoch * epochs` batches (in this case, 28100 batches). You may need to use the repeat() function when building your dataset.
WARNING:tensorflow:Your input ran out of data; interrupting training. Make sure that your dataset or generator can generate at least `steps_per_epoch * epochs` batches (in this case, 28100 batches). You may need to use the repeat() function when building your dataset.
562/562 [==============================] - 23s 42ms/step - loss: 0.7348 - accuracy: 0.7100 - f1: 0.6739 - val_loss: 0.6214 - val_accuracy: 0.7638 - val_f1: 0.7549
Learning Curve¶
plot_learning_curve(model, extra_metric='accuracy');
Predictions¶
# Make predictions
PREDICTIONS[model_name] = {}
for subset, dset in ds.items():
PREDICTIONS[model_name][subset] = reshape_classification_prediction(model.predict(dset))
Model 5: Transfer Learning with pretrained sentence embeddings (USE) + character embeddings + positional embeddings¶
- Combining the embeddings lead to a slightly better performance.
- But there is one major information that is missing about the sentence. If we notice, it is always the case that the order of the categories is fixed i.e.
OBJECTIVE->METHODS->RESULTS->CONCLUSIONS - When we parsed the structure of the data, we also added the line_number and total_lines to the parsed dictionary for each sample.
data_samples['train'].head()
| id | target | text | line_number | total_lines | |
|---|---|---|---|---|---|
| 0 | 24293578 | OBJECTIVE | To investigate the efficacy of @ weeks of dail... | 0 | 11 |
| 1 | 24293578 | METHODS | A total of @ patients with primary knee OA wer... | 1 | 11 |
| 2 | 24293578 | METHODS | Outcome measures included pain reduction and i... | 2 | 11 |
| 3 | 24293578 | METHODS | Pain was assessed using the visual analog pain... | 3 | 11 |
| 4 | 24293578 | METHODS | Secondary outcome measures included the Wester... | 4 | 11 |
# Value counts of line_numbers
plt.figure(figsize=(12, 4))
data_samples['train']['line_number'].value_counts().sort_index().plot(kind='bar');
(data_samples['train']['line_number'] > 15).mean()*100
1.7223950233281493
Majority of the lines have a position of 15 or less (only 1.72 % has more than 15 line number). Hence 15 is probably the 98th percentile. We can featurize this by one-hot-encoding the positions and limiting the depth to be less than 15.
line_numbers_features = {}
for subset in ['train', 'dev', 'test']:
line_numbers_features[subset] = tf.one_hot(data_samples[subset]['line_number'], depth=15)
Let us do the same for total_lines feature
# Value counts of total_lines
plt.figure(figsize=(12, 4))
data_samples['train']['total_lines'].value_counts().sort_index().plot(kind='bar');
Let us check the 98th percentile (similar to line_number), and we can limit the maximum total_lines accordingly
max_total_lines = np.percentile(data_samples['train']['total_lines'], 98)
max_total_lines
20.0
Great! Now let us one-hot-encode the total_lines by limiting the max depth to 20
total_lines_features = {}
for subset in ['train', 'dev', 'test']:
total_lines_features[subset] = tf.one_hot(data_samples[subset]['total_lines'], depth=20)
Building the hybrid three embedding model¶
We are going to build a model which combines the USE, Character level, and Positional embeddings
These are the steps we are going to be following:
- Create the USE embedding model using sentences as input
- Create the *character embedding model using character (sentence with extra space) as inputs
- Create line_number model which takes in one-hot encoded
line_numbertensors, passes through some non-linear layers and learns an encoding for each line_number- Alternatively, we could have done it by passing integer encoded line_numbers, and passed it through an embedding layer, and this would have been a similar, as one-hot encoded vector + a non linear layer.
- Create a total_lines model which takes in one-hot encoded
total_linestensors, passes through some non-linear layers and learns an encoded for each total_lines - Combine the outputs of 1. (USE) and 2. (character) by stacking/concatenating them and pass them through some non linear layers.
- Combine the outputs of 3. (line_number), 4. (total_lines) and 5. by concatenating/stacking them
- Now create the final output layer with N_CLASSES, and predict the softmax probabilities.
../re

LINE_NUMBERS_FEATURE_SHAPE = line_numbers_features['train'].shape[1]
TOTAL_LINES_FEATURE_SHAPE = total_lines_features['train'].shape[1]
model_name = 'use-char-pos-embed-tribrid'
# 1. USE embeddings model
use_model_inputs = layers.Input(shape=[], dtype=tf.string)
sent_embeddings = use_embed_layer(use_model_inputs)
use_model_outputs = layers.Dense(128, activation='relu')(sent_embeddings)
use_model = tf.keras.Model(use_model_inputs, use_model_outputs, name='use_embed_model')
# 2. Character embeddings model
char_model_inputs = layers.Input(shape=(1,), dtype=tf.string)
char_vec = char_vectorizer(char_model_inputs)
char_embedding = char_embed(char_vec)
char_bi_lstm = layers.Bidirectional(layers.LSTM(32))(char_embedding)
char_model = tf.keras.Model(char_model_inputs, char_bi_lstm, name='char_embed_model')
# 3. Line number model
line_number_inputs = layers.Input(shape=LINE_NUMBERS_FEATURE_SHAPE)
line_number_outputs = layers.Dense(32, activation='relu')(line_number_inputs)
line_number_model = tf.keras.Model(inputs=line_number_inputs, outputs=line_number_outputs,
name='line_number_model')
# 4. Total lines model
total_lines_input = layers.Input(shape=TOTAL_LINES_FEATURE_SHAPE)
total_lines_output = layers.Dense(32, activation='relu')(total_lines_input)
total_lines_model = tf.keras.Model(inputs=total_lines_input, outputs=total_lines_output,
name='total_lines_model')
# 5. Combined embedding of USE (sentence) and character
comb_embeddings_sent_char = layers.Concatenate(name='sent_char_hybrid_embedding')([use_model.output, char_model.output])
comb_embeddings_sent_char_repr = layers.Dense(256, activation='relu')(comb_embeddings_sent_char)
comb_embeddings_sent_char_repr = layers.Dropout(0.5)(comb_embeddings_sent_char_repr)
# 6. Combine embedding repr output of 5., with 3. line_number repr output and 4. total_lines repr output
comb_embeddings_all = layers.Concatenate(name='combined_embeddings_all')([line_number_model.output,
total_lines_model.output,
comb_embeddings_sent_char_repr])
# 7. Final output layer which predicts the softmax probabilities
output_layer = layers.Dense(N_CLASSES, activation='softmax', name='output_layer')(comb_embeddings_all)
# Put together the whole model
model = tf.keras.Model(inputs=[use_model.input, char_model.input,
line_number_model.input, total_lines_model.input],
outputs=output_layer, name=model_name)
model.summary()
Model: "use-char-pos-embed-tribrid"
__________________________________________________________________________________________________
Layer (type) Output Shape Param # Connected to
==================================================================================================
input_66 (InputLayer) [(None, 1)] 0
__________________________________________________________________________________________________
input_65 (InputLayer) [(None,)] 0
__________________________________________________________________________________________________
char_vectorizer (TextVectorizat (None, 290) 0 input_66[0][0]
__________________________________________________________________________________________________
universal_sentence_encoder (Ker (None, 512) 256797824 input_65[0][0]
__________________________________________________________________________________________________
char_embed (Embedding) (None, 290, 25) 700 char_vectorizer[2][0]
__________________________________________________________________________________________________
dense_63 (Dense) (None, 128) 65664 universal_sentence_encoder[2][0]
__________________________________________________________________________________________________
bidirectional_13 (Bidirectional (None, 64) 14848 char_embed[2][0]
__________________________________________________________________________________________________
sent_char_hybrid_embedding (Con (None, 192) 0 dense_63[0][0]
bidirectional_13[0][0]
__________________________________________________________________________________________________
input_67 (InputLayer) [(None, 15)] 0
__________________________________________________________________________________________________
input_68 (InputLayer) [(None, 20)] 0
__________________________________________________________________________________________________
dense_66 (Dense) (None, 256) 49408 sent_char_hybrid_embedding[0][0]
__________________________________________________________________________________________________
dense_64 (Dense) (None, 32) 512 input_67[0][0]
__________________________________________________________________________________________________
dense_65 (Dense) (None, 32) 672 input_68[0][0]
__________________________________________________________________________________________________
dropout_12 (Dropout) (None, 256) 0 dense_66[0][0]
__________________________________________________________________________________________________
combined_embeddings_all (Concat (None, 320) 0 dense_64[0][0]
dense_65[0][0]
dropout_12[0][0]
__________________________________________________________________________________________________
output_layer (Dense) (None, 5) 1605 combined_embeddings_all[0][0]
==================================================================================================
Total params: 256,931,233
Trainable params: 133,409
Non-trainable params: 256,797,824
__________________________________________________________________________________________________
Plot the architecture of the model¶
tf.keras.utils.plot_model(model, to_file='/tmp/model.png', show_shapes=True)
The general premise of embeddings, and passing features through hidden layers is to learn appropriate numerical representation of each i.e. encoding information in a way that is useful for the model to achieve its supervised task.
Our model is very similar to the paper Figure 1 of Neural Networks for Joint Sentence Classification in Medical Paper Abstracts. But in a few places, its a bit different:
- We are using Universal Sentence Encoder instead of GloVe embeddings.
- To use the GloVe embeddings, we have to download the vectors for a vocabulary (various sizes) -> create an embedding matrix for our vocab -> initialize the embedding layer with that.
- We are passing the Sentence level embedding through a Dense layer, and passing the character level embedding through the Bi-LSTM
- Section 3.1.3 of the paper uses a label optimization layer (which makes the sure sequence labels come out in a specific order)
- This is very similar to Conditional Random Fields, and will help prevent overfitting.
- We used the postional embedding (sort of) by encoding the information of line_number and total_lines, as a replacement of this layer.
- Paper in Section 4.2 mentions that they update the character and token embeddings (in our case sentence embeddings). We keep the USE layer frozen, and only update the character level embedding.
- Paper uses
StochasticGradientDescent, and we use the old and trustAdamoptimizer.
We are going to modify the CategoricalCrossEntropy loss function just a little bit by introducing a parameter called label_smoothing, which will help reduce overfitting and improve generalization of the result.
This is somewhat similar to the concept of Temperature Scaling of probabilities.
For example instead of having a totally overconfident prediction of
[0.0, 1.0, 0.0, 0.0, 0.0]
We smooth out the predictions just a little bit as follows:
[0.01, 0.96, 0.01, 0.01, 0.01]
Resource: PyImageSearch: Label smoothing with Keras, TensorFlow, and Deep Learning.
model.compile(loss=tf.keras.losses.CategoricalCrossentropy(label_smoothing=0.2),
optimizer=tf.keras.optimizers.Adam(), metrics=['accuracy', KerasMetrics.f1])
Creating a specific tf.data.Dataset for this model¶
Again, stressing, that using tf.data.Dataset API will help in more efficiently passing the inputs to the model through mapping a function, prefetching and batching (also parellization)
Note: Pass the datasets in a tuple and NOT in a list. Weird bug.
sent_char_pos_tribrid_dataset = {}
for subset in ['train', 'dev', 'test']:
tfdata_inputs = tf.data.Dataset.from_tensor_slices((sentences[subset], characters[subset],
line_numbers_features[subset], total_lines_features[subset]))
tfdata_labels = tf.data.Dataset.from_tensor_slices(data_labels['one_hot'][subset])
tfdata = tf.data.Dataset.zip((tfdata_inputs, tfdata_labels)).batch(32).prefetch(buffer_size=tf.data.AUTOTUNE)
sent_char_pos_tribrid_dataset[subset] = tfdata
Now finally, FIT the model¶
ds = sent_char_pos_tribrid_dataset
train_steps = int(0.1*len(ds['train']))
val_steps = int(0.1*len(ds['dev']))
model.fit(ds['train'], steps_per_epoch=train_steps,
validation_data=ds['dev'], validation_steps=val_steps,
epochs=NUM_EPOCHS)
MODELS[model_name] = model
Epoch 1/50 562/562 [==============================] - 47s 67ms/step - loss: 1.0925 - accuracy: 0.7323 - f1: 0.6153 - val_loss: 0.9817 - val_accuracy: 0.8049 - val_f1: 0.7485 Epoch 2/50 562/562 [==============================] - 35s 62ms/step - loss: 0.9674 - accuracy: 0.8159 - f1: 0.7685 - val_loss: 0.9490 - val_accuracy: 0.8324 - val_f1: 0.7809 Epoch 3/50 562/562 [==============================] - 31s 55ms/step - loss: 0.9508 - accuracy: 0.8248 - f1: 0.7817 - val_loss: 0.9412 - val_accuracy: 0.8291 - val_f1: 0.7934 Epoch 4/50 562/562 [==============================] - 36s 64ms/step - loss: 0.9399 - accuracy: 0.8339 - f1: 0.8000 - val_loss: 0.9291 - val_accuracy: 0.8381 - val_f1: 0.8119 Epoch 5/50 562/562 [==============================] - 33s 58ms/step - loss: 0.9362 - accuracy: 0.8373 - f1: 0.8042 - val_loss: 0.9220 - val_accuracy: 0.8394 - val_f1: 0.8158 Epoch 6/50 562/562 [==============================] - 32s 57ms/step - loss: 0.9396 - accuracy: 0.8304 - f1: 0.7970 - val_loss: 0.9182 - val_accuracy: 0.8441 - val_f1: 0.8169 Epoch 7/50 562/562 [==============================] - 38s 68ms/step - loss: 0.9280 - accuracy: 0.8411 - f1: 0.8094 - val_loss: 0.9184 - val_accuracy: 0.8447 - val_f1: 0.8182 Epoch 8/50 562/562 [==============================] - 35s 62ms/step - loss: 0.9228 - accuracy: 0.8435 - f1: 0.8145 - val_loss: 0.9063 - val_accuracy: 0.8570 - val_f1: 0.8235 Epoch 9/50 562/562 [==============================] - 34s 61ms/step - loss: 0.9249 - accuracy: 0.8421 - f1: 0.8138 - val_loss: 0.9178 - val_accuracy: 0.8418 - val_f1: 0.8272 Epoch 10/50 562/562 [==============================] - 36s 64ms/step - loss: 0.9216 - accuracy: 0.8454 - f1: 0.8156 - val_loss: 0.9089 - val_accuracy: 0.8431 - val_f1: 0.8233 Epoch 11/50 7/562 [..............................] - ETA: 25s - loss: 0.9308 - accuracy: 0.8350 - f1: 0.7818WARNING:tensorflow:Your input ran out of data; interrupting training. Make sure that your dataset or generator can generate at least `steps_per_epoch * epochs` batches (in this case, 28100 batches). You may need to use the repeat() function when building your dataset.
WARNING:tensorflow:Your input ran out of data; interrupting training. Make sure that your dataset or generator can generate at least `steps_per_epoch * epochs` batches (in this case, 28100 batches). You may need to use the repeat() function when building your dataset.
562/562 [==============================] - 3s 5ms/step - loss: 0.9308 - accuracy: 0.8350 - f1: 0.7818 - val_loss: 0.9136 - val_accuracy: 0.8414 - val_f1: 0.8189
Learning Curve¶
plot_learning_curve(model, extra_metric='accuracy');
# Make predictions
PREDICTIONS[model_name] = {}
for subset, dset in ds.items():
PREDICTIONS[model_name][subset] = reshape_classification_prediction(model.predict(dset))
Save the models¶
import pickle
for model_name, model in MODELS.items():
model_savename = f'../models/pubmed_rct_abstract_multiclass_classification/{model_name}'
if model_name == 'naive-bayes-baseline':
if not os.path.exists(model_savename):
os.makedirs(model_savename)
with open(f'{model_savename}/{model_name}.pkl', 'wb') as f:
pickle.dump(model, f)
continue
model.save(model_savename)
INFO:tensorflow:Assets written to: ../models/pubmed_rct_abstract_multiclass_classification/Conv1D-word-embed/assets
INFO:tensorflow:Assets written to: ../models/pubmed_rct_abstract_multiclass_classification/Conv1D-word-embed/assets
INFO:tensorflow:Assets written to: ../models/pubmed_rct_abstract_multiclass_classification/USE-feature-extraction/assets
INFO:tensorflow:Assets written to: ../models/pubmed_rct_abstract_multiclass_classification/USE-feature-extraction/assets
INFO:tensorflow:Assets written to: ../models/pubmed_rct_abstract_multiclass_classification/Conv1D-char-embed/assets
INFO:tensorflow:Assets written to: ../models/pubmed_rct_abstract_multiclass_classification/Conv1D-char-embed/assets WARNING:absl:Found untraced functions such as lstm_cell_37_layer_call_and_return_conditional_losses, lstm_cell_37_layer_call_fn, lstm_cell_38_layer_call_and_return_conditional_losses, lstm_cell_38_layer_call_fn, lstm_cell_37_layer_call_fn while saving (showing 5 of 10). These functions will not be directly callable after loading.
INFO:tensorflow:Assets written to: ../models/pubmed_rct_abstract_multiclass_classification/USE-char-hybrid-embed/assets
INFO:tensorflow:Assets written to: ../models/pubmed_rct_abstract_multiclass_classification/USE-char-hybrid-embed/assets WARNING:absl:Found untraced functions such as lstm_cell_40_layer_call_and_return_conditional_losses, lstm_cell_40_layer_call_fn, lstm_cell_41_layer_call_and_return_conditional_losses, lstm_cell_41_layer_call_fn, lstm_cell_40_layer_call_fn while saving (showing 5 of 10). These functions will not be directly callable after loading.
INFO:tensorflow:Assets written to: ../models/pubmed_rct_abstract_multiclass_classification/use-char-pos-embed-tribrid/assets
INFO:tensorflow:Assets written to: ../models/pubmed_rct_abstract_multiclass_classification/use-char-pos-embed-tribrid/assets
Save Predictions¶
with open('../scratch/predictions.pkl', 'wb') as f:
pickle.dump(PREDICTIONS, f)
Performance Comparison¶
from src.evaluate import ClassificationPerformanceComparer
data_labels['label'][subset].reshape(-1, 1)
array([[0],
[4],
[4],
...,
[4],
[4],
[1]])
clf_comps = {}
for subset in ['train', 'dev', 'test']:
predictions = [pred_dict[subset] for pred_dict in PREDICTIONS.values()]
clf_comps[subset] = ClassificationPerformanceComparer.from_predictions(
predictions=predictions, data=(None, data_labels['label'][subset].reshape(-1, 1)),
model_names=list(MODELS.keys()), class_names=CLASS_NAMES)
fig, axn = plt.subplots(3, 1, figsize=(12, 12))
for ax, (subset, clf_comp) in zip(axn, clf_comps.items()):
clf_comp.calculate_metric_comparison_df()
clf_comp.plot_metric_comparison_df(ax=ax)
ax.set_title(subset, fontdict=dict(weight='bold', size=20))
handles, labels = ax.get_legend_handles_labels()
ax.get_legend().remove()
fig.legend(handles, labels, loc='center right', bbox_to_anchor=(1.05, 0.5))
plt.tight_layout()
WARNING:matplotlib.legend:No handles with labels found to put in legend. WARNING:matplotlib.legend:No handles with labels found to put in legend.
The paper PubMed 200k RCT:
a Dataset for Sequential Sentence Classification in Medical Abstracts compared the F1-score of the models (they achieved 90.0%). Our best model use-char-pos-embed-tribrid of around 84%, but we only trained for 10 epochs and on only 10% of even the 20k RCT data.
Finding the most wrong predictions¶
Most wrong predictions are those which are wrong (duh!) and predicted with a high confidence.
import pickle
with open('../scratch/predictions.pkl', 'rb') as f:
PREDICTIONS = pickle.load(f)
PREDICTIONS.keys()
dict_keys(['naive-bayes-baseline', 'Conv1D-word-embed', 'USE-feature-extraction', 'Conv1D-char-embed', 'USE-char-hybrid-embed', 'use-char-pos-embed-tribrid'])
CLASS_NAMES
['OBJECTIVE', 'METHODS', 'RESULTS', 'CONCLUSIONS', 'BACKGROUND']
subset = 'test'
model_name = 'use-char-pos-embed-tribrid'
df = data_samples[subset]
df['label'] = data_labels['label'][subset]
df['pred'] = PREDICTIONS[model_name][subset].argmax(axis=1)
df['pred_prob'] = PREDICTIONS[model_name][subset].max(axis=1)
df['pred_name'] = df['pred'].map({i: name for i, name in enumerate(CLASS_NAMES)})
df = df[['id', 'text', 'line_number', 'total_lines', 'target',
'label', 'pred_name', 'pred', 'pred_prob']]
df.head()
| id | text | line_number | total_lines | target | label | pred_name | pred | pred_prob | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 24845963 | This study analyzed liver function abnormaliti... | 0 | 8 | BACKGROUND | 0 | BACKGROUND | 0 | 0.421630 |
| 1 | 24845963 | A post hoc analysis was conducted with the use... | 1 | 8 | RESULTS | 4 | METHODS | 2 | 0.466157 |
| 2 | 24845963 | Liver function tests ( LFTs ) were measured at... | 2 | 8 | RESULTS | 4 | METHODS | 2 | 0.833757 |
| 3 | 24845963 | Survival analyses were used to assess the asso... | 3 | 8 | RESULTS | 4 | METHODS | 2 | 0.799859 |
| 4 | 24845963 | The percentage of patients with abnormal LFTs ... | 4 | 8 | RESULTS | 4 | RESULTS | 4 | 0.777786 |
most_wrong = df.loc[df['label'] != df['pred']].sort_values('pred_prob', ascending=False).reset_index(drop=True)
most_wrong
| id | text | line_number | total_lines | target | label | pred_name | pred | pred_prob | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 25036218 | NCT@ ( ClinicalTrials.gov ) . | 18 | 18 | CONCLUSIONS | 1 | BACKGROUND | 0 | 0.958825 |
| 1 | 25204768 | Pretest-posttest . | 1 | 11 | METHODS | 2 | BACKGROUND | 0 | 0.951528 |
| 2 | 24786163 | Of the @ dogs receiving placebo , @ ( @ % ) vo... | 7 | 10 | METHODS | 2 | RESULTS | 4 | 0.936474 |
| 3 | 25587059 | non-diffuse-trickling '' ) . | 8 | 15 | RESULTS | 4 | BACKGROUND | 0 | 0.935319 |
| 4 | 24886555 | Symptom outcomes will be assessed and estimate... | 4 | 6 | CONCLUSIONS | 1 | METHODS | 2 | 0.932493 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 4753 | 24625034 | These data will allow the development of a lar... | 7 | 16 | METHODS | 2 | RESULTS | 4 | 0.252344 |
| 4754 | 25190617 | The objective of the IDEAL DVT study , therefo... | 6 | 16 | BACKGROUND | 0 | RESULTS | 4 | 0.250690 |
| 4755 | 25194287 | Results from smaller prospective , randomized ... | 3 | 9 | BACKGROUND | 0 | CONCLUSIONS | 1 | 0.247399 |
| 4756 | 25522560 | The therapeutic effect of macrolides was also ... | 2 | 6 | METHODS | 2 | BACKGROUND | 0 | 0.240238 |
| 4757 | 24245807 | The aim of the present study was to investigat... | 4 | 11 | BACKGROUND | 0 | RESULTS | 4 | 0.240217 |
4758 rows × 9 columns
for i, row in most_wrong[0:10].iterrows():
print('Target: {target}, Pred: {pred_name}, Prob: {pred_prob:.2f}\n'.format(**row))
print('Sentence:\n{}\n'.format(row['text']))
print('----\n')
Target: CONCLUSIONS, Pred: BACKGROUND, Prob: 0.96 Sentence: NCT@ ( ClinicalTrials.gov ) . ---- Target: METHODS, Pred: BACKGROUND, Prob: 0.95 Sentence: Pretest-posttest . ---- Target: METHODS, Pred: RESULTS, Prob: 0.94 Sentence: Of the @ dogs receiving placebo , @ ( @ % ) vomited and @ ( @ % ) developed signs of nausea ; overall , @ of @ ( @ % ) dogs in the placebo treatment group vomited or developed signs of nausea . ---- Target: RESULTS, Pred: BACKGROUND, Prob: 0.94 Sentence: non-diffuse-trickling '' ) . ---- Target: CONCLUSIONS, Pred: METHODS, Prob: 0.93 Sentence: Symptom outcomes will be assessed and estimates of cost-effectiveness made . ---- Target: BACKGROUND, Pred: OBJECTIVE, Prob: 0.92 Sentence: To evaluate the effects of the lactic acid bacterium Lactobacillus salivarius on caries risk factors . ---- Target: OBJECTIVE, Pred: METHODS, Prob: 0.92 Sentence: The National Institute of Oncology , Budapest conducted a single centre randomized clinical study . ---- Target: RESULTS, Pred: METHODS, Prob: 0.92 Sentence: Baseline measures included sociodemographics , standardized anthropometrics , Asthma Control Test ( ACT ) , GERD Symptom Assessment Scale , Pittsburgh Sleep Quality Index , and Berlin Questionnaire for Sleep Apnea . ---- Target: RESULTS, Pred: METHODS, Prob: 0.92 Sentence: A cluster randomised trial was implemented with @,@ children in @ government primary schools on the south coast of Kenya in @-@ . ---- Target: RESULTS, Pred: METHODS, Prob: 0.91 Sentence: The primary endpoint is the cumulative three-year HIV incidence . ----
Some of the examples seem like they would end up confusing the model more, or they maybe ambiguous. We need to avoid passing such instances to the model for learning (or maybe penalise them less for ambiguous samples?). Or we may need to identify wrongly labelled samples, and relabel them, and then retrain the model. This is called Active Learning or Human in the loop learning
Make example predictions¶
!wget https://raw.githubusercontent.com/mrdbourke/tensorflow-deep-learning/main/extras/skimlit_example_abstracts.json ../data/
--2021-06-21 17:32:06-- https://raw.githubusercontent.com/mrdbourke/tensorflow-deep-learning/main/extras/skimlit_example_abstracts.json Resolving raw.githubusercontent.com (raw.githubusercontent.com)... 185.199.108.133, 185.199.109.133, 185.199.110.133, ... Connecting to raw.githubusercontent.com (raw.githubusercontent.com)|185.199.108.133|:443... connected. HTTP request sent, awaiting response... 200 OK Length: 6737 (6.6K) [text/plain] Saving to: ‘skimlit_example_abstracts.json’ skimlit_example_abs 100%[===================>] 6.58K --.-KB/s in 0.001s 2021-06-21 17:32:06 (5.79 MB/s) - ‘skimlit_example_abstracts.json’ saved [6737/6737] --2021-06-21 17:32:06-- http://../data/ Resolving .. (..)... failed: Name or service not known. wget: unable to resolve host address ‘..’ FINISHED --2021-06-21 17:32:06-- Total wall clock time: 0.2s Downloaded: 1 files, 6.6K in 0.001s (5.79 MB/s)
import json
with open('skimlit_example_abstracts.json', 'r') as f:
example_abstracts = json.load(f)
example_abstracts
[{'abstract': 'This RCT examined the efficacy of a manualized social intervention for children with HFASDs. Participants were randomly assigned to treatment or wait-list conditions. Treatment included instruction and therapeutic activities targeting social skills, face-emotion recognition, interest expansion, and interpretation of non-literal language. A response-cost program was applied to reduce problem behaviors and foster skills acquisition. Significant treatment effects were found for five of seven primary outcome measures (parent ratings and direct child measures). Secondary measures based on staff ratings (treatment group only) corroborated gains reported by parents. High levels of parent, child and staff satisfaction were reported, along with high levels of treatment fidelity. Standardized effect size estimates were primarily in the medium and large ranges and favored the treatment group.',
'details': 'RCT of a manualized social treatment for high-functioning autism spectrum disorders',
'source': 'https://pubmed.ncbi.nlm.nih.gov/20232240/'},
{'abstract': "Postpartum depression (PPD) is the most prevalent mood disorder associated with childbirth. No single cause of PPD has been identified, however the increased risk of nutritional deficiencies incurred through the high nutritional requirements of pregnancy may play a role in the pathology of depressive symptoms. Three nutritional interventions have drawn particular interest as possible non-invasive and cost-effective prevention and/or treatment strategies for PPD; omega-3 (n-3) long chain polyunsaturated fatty acids (LCPUFA), vitamin D and overall diet. We searched for meta-analyses of randomised controlled trials (RCT's) of nutritional interventions during the perinatal period with PPD as an outcome, and checked for any trials published subsequently to the meta-analyses. Fish oil: Eleven RCT's of prenatal fish oil supplementation RCT's show null and positive effects on PPD symptoms. Vitamin D: no relevant RCT's were identified, however seven observational studies of maternal vitamin D levels with PPD outcomes showed inconsistent associations. Diet: Two Australian RCT's with dietary advice interventions in pregnancy had a positive and null result on PPD. With the exception of fish oil, few RCT's with nutritional interventions during pregnancy assess PPD. Further research is needed to determine whether nutritional intervention strategies during pregnancy can protect against symptoms of PPD. Given the prevalence of PPD and ease of administering PPD measures, we recommend future prenatal nutritional RCT's include PPD as an outcome.",
'details': 'Formatting removed (can be used to compare model to actual example)',
'source': 'https://pubmed.ncbi.nlm.nih.gov/28012571/'},
{'abstract': 'Mental illness, including depression, anxiety and bipolar disorder, accounts for a significant proportion of global disability and poses a substantial social, economic and heath burden. Treatment is presently dominated by pharmacotherapy, such as antidepressants, and psychotherapy, such as cognitive behavioural therapy; however, such treatments avert less than half of the disease burden, suggesting that additional strategies are needed to prevent and treat mental disorders. There are now consistent mechanistic, observational and interventional data to suggest diet quality may be a modifiable risk factor for mental illness. This review provides an overview of the nutritional psychiatry field. It includes a discussion of the neurobiological mechanisms likely modulated by diet, the use of dietary and nutraceutical interventions in mental disorders, and recommendations for further research. Potential biological pathways related to mental disorders include inflammation, oxidative stress, the gut microbiome, epigenetic modifications and neuroplasticity. Consistent epidemiological evidence, particularly for depression, suggests an association between measures of diet quality and mental health, across multiple populations and age groups; these do not appear to be explained by other demographic, lifestyle factors or reverse causality. Our recently published intervention trial provides preliminary clinical evidence that dietary interventions in clinically diagnosed populations are feasible and can provide significant clinical benefit. Furthermore, nutraceuticals including n-3 fatty acids, folate, S-adenosylmethionine, N-acetyl cysteine and probiotics, among others, are promising avenues for future research. Continued research is now required to investigate the efficacy of intervention studies in large cohorts and within clinically relevant populations, particularly in patients with schizophrenia, bipolar and anxiety disorders.',
'details': 'Effect of nutrition on mental health',
'source': 'https://pubmed.ncbi.nlm.nih.gov/28942748/'},
{'abstract': "Hepatitis C virus (HCV) and alcoholic liver disease (ALD), either alone or in combination, count for more than two thirds of all liver diseases in the Western world. There is no safe level of drinking in HCV-infected patients and the most effective goal for these patients is total abstinence. Baclofen, a GABA(B) receptor agonist, represents a promising pharmacotherapy for alcohol dependence (AD). Previously, we performed a randomized clinical trial (RCT), which demonstrated the safety and efficacy of baclofen in patients affected by AD and cirrhosis. The goal of this post-hoc analysis was to explore baclofen's effect in a subgroup of alcohol-dependent HCV-infected cirrhotic patients. Any patient with HCV infection was selected for this analysis. Among the 84 subjects randomized in the main trial, 24 alcohol-dependent cirrhotic patients had a HCV infection; 12 received baclofen 10mg t.i.d. and 12 received placebo for 12-weeks. With respect to the placebo group (3/12, 25.0%), a significantly higher number of patients who achieved and maintained total alcohol abstinence was found in the baclofen group (10/12, 83.3%; p=0.0123). Furthermore, in the baclofen group, compared to placebo, there was a significantly higher increase in albumin values from baseline (p=0.0132) and a trend toward a significant reduction in INR levels from baseline (p=0.0716). In conclusion, baclofen was safe and significantly more effective than placebo in promoting alcohol abstinence, and improving some Liver Function Tests (LFTs) (i.e. albumin, INR) in alcohol-dependent HCV-infected cirrhotic patients. Baclofen may represent a clinically relevant alcohol pharmacotherapy for these patients.",
'details': 'Baclofen promotes alcohol abstinence in alcohol dependent cirrhotic patients with hepatitis C virus (HCV) infection',
'source': 'https://pubmed.ncbi.nlm.nih.gov/22244707/'}]
abstract_df = pd.DataFrame(example_abstracts)
abstract_df
| abstract | source | details | |
|---|---|---|---|
| 0 | This RCT examined the efficacy of a manualized... | https://pubmed.ncbi.nlm.nih.gov/20232240/ | RCT of a manualized social treatment for high-... |
| 1 | Postpartum depression (PPD) is the most preval... | https://pubmed.ncbi.nlm.nih.gov/28012571/ | Formatting removed (can be used to compare mod... |
| 2 | Mental illness, including depression, anxiety ... | https://pubmed.ncbi.nlm.nih.gov/28942748/ | Effect of nutrition on mental health |
| 3 | Hepatitis C virus (HCV) and alcoholic liver di... | https://pubmed.ncbi.nlm.nih.gov/22244707/ | Baclofen promotes alcohol abstinence in alcoho... |
- We need to break the abstract into proper sentences. Just splitting on full stop
.won't help. We need a proper sentence parser. Enterspacysentencizer - Then for each sentences we need to extract the
line_numberandtotal_linesfeature. This we need to one-hot-encode. - The other two inputs to the model are the sentence itself (which will be passed to USE model), and the sentence with extra space between each character (which will be passed Character model)
from spacy.lang.en import English
nlp = English()
sentencizer = nlp.create_pipe('sentencizer')
nlp.add_pipe(sentencizer)
doc = nlp(example_abstracts[0]['abstract'])
abstract_lines = [str(sent) for sent in list(doc.sents)]
abstract_lines
['This RCT examined the efficacy of a manualized social intervention for children with HFASDs.', 'Participants were randomly assigned to treatment or wait-list conditions.', 'Treatment included instruction and therapeutic activities targeting social skills, face-emotion recognition, interest expansion, and interpretation of non-literal language.', 'A response-cost program was applied to reduce problem behaviors and foster skills acquisition.', 'Significant treatment effects were found for five of seven primary outcome measures (parent ratings and direct child measures).', 'Secondary measures based on staff ratings (treatment group only) corroborated gains reported by parents.', 'High levels of parent, child and staff satisfaction were reported, along with high levels of treatment fidelity.', 'Standardized effect size estimates were primarily in the medium and large ranges and favored the treatment group.']
total_lines = len(abstract_lines)
sample_lines = []
for i, line in enumerate(abstract_lines):
sample_dict = {}
sample_dict['text'] = line
sample_dict['line_number'] = i
sample_dict['total_lines'] = total_lines
sample_lines.append(sample_dict)
sample_lines
[{'line_number': 0,
'text': 'This RCT examined the efficacy of a manualized social intervention for children with HFASDs.',
'total_lines': 8},
{'line_number': 1,
'text': 'Participants were randomly assigned to treatment or wait-list conditions.',
'total_lines': 8},
{'line_number': 2,
'text': 'Treatment included instruction and therapeutic activities targeting social skills, face-emotion recognition, interest expansion, and interpretation of non-literal language.',
'total_lines': 8},
{'line_number': 3,
'text': 'A response-cost program was applied to reduce problem behaviors and foster skills acquisition.',
'total_lines': 8},
{'line_number': 4,
'text': 'Significant treatment effects were found for five of seven primary outcome measures (parent ratings and direct child measures).',
'total_lines': 8},
{'line_number': 5,
'text': 'Secondary measures based on staff ratings (treatment group only) corroborated gains reported by parents.',
'total_lines': 8},
{'line_number': 6,
'text': 'High levels of parent, child and staff satisfaction were reported, along with high levels of treatment fidelity.',
'total_lines': 8},
{'line_number': 7,
'text': 'Standardized effect size estimates were primarily in the medium and large ranges and favored the treatment group.',
'total_lines': 8}]
abstract_line_numbers_one_hot = tf.one_hot([line['line_number'] for line in sample_lines], depth=15)
abstract_line_numbers_one_hot
<tf.Tensor: shape=(8, 15), dtype=float32, numpy=
array([[1., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0.],
[0., 1., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0.],
[0., 0., 1., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0.],
[0., 0., 0., 1., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0.],
[0., 0., 0., 0., 1., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0., 1., 0., 0., 0., 0., 0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0., 0., 1., 0., 0., 0., 0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0., 0., 0., 1., 0., 0., 0., 0., 0., 0., 0.]],
dtype=float32)>
abstract_total_lines_one_hot = tf.one_hot([line['total_lines'] for line in sample_lines], depth=20)
abstract_total_lines_one_hot
<tf.Tensor: shape=(8, 20), dtype=float32, numpy=
array([[0., 0., 0., 0., 0., 0., 0., 0., 1., 0., 0., 0., 0., 0., 0., 0.,
0., 0., 0., 0.],
[0., 0., 0., 0., 0., 0., 0., 0., 1., 0., 0., 0., 0., 0., 0., 0.,
0., 0., 0., 0.],
[0., 0., 0., 0., 0., 0., 0., 0., 1., 0., 0., 0., 0., 0., 0., 0.,
0., 0., 0., 0.],
[0., 0., 0., 0., 0., 0., 0., 0., 1., 0., 0., 0., 0., 0., 0., 0.,
0., 0., 0., 0.],
[0., 0., 0., 0., 0., 0., 0., 0., 1., 0., 0., 0., 0., 0., 0., 0.,
0., 0., 0., 0.],
[0., 0., 0., 0., 0., 0., 0., 0., 1., 0., 0., 0., 0., 0., 0., 0.,
0., 0., 0., 0.],
[0., 0., 0., 0., 0., 0., 0., 0., 1., 0., 0., 0., 0., 0., 0., 0.,
0., 0., 0., 0.],
[0., 0., 0., 0., 0., 0., 0., 0., 1., 0., 0., 0., 0., 0., 0., 0.,
0., 0., 0., 0.]], dtype=float32)>
abstract_chars = [split_chars(sent, return_string=True) for sent in abstract_lines]
abstract_chars
['T h i s R C T e x a m i n e d t h e e f f i c a c y o f a m a n u a l i z e d s o c i a l i n t e r v e n t i o n f o r c h i l d r e n w i t h H F A S D s .', 'P a r t i c i p a n t s w e r e r a n d o m l y a s s i g n e d t o t r e a t m e n t o r w a i t - l i s t c o n d i t i o n s .', 'T r e a t m e n t i n c l u d e d i n s t r u c t i o n a n d t h e r a p e u t i c a c t i v i t i e s t a r g e t i n g s o c i a l s k i l l s , f a c e - e m o t i o n r e c o g n i t i o n , i n t e r e s t e x p a n s i o n , a n d i n t e r p r e t a t i o n o f n o n - l i t e r a l l a n g u a g e .', 'A r e s p o n s e - c o s t p r o g r a m w a s a p p l i e d t o r e d u c e p r o b l e m b e h a v i o r s a n d f o s t e r s k i l l s a c q u i s i t i o n .', 'S i g n i f i c a n t t r e a t m e n t e f f e c t s w e r e f o u n d f o r f i v e o f s e v e n p r i m a r y o u t c o m e m e a s u r e s ( p a r e n t r a t i n g s a n d d i r e c t c h i l d m e a s u r e s ) .', 'S e c o n d a r y m e a s u r e s b a s e d o n s t a f f r a t i n g s ( t r e a t m e n t g r o u p o n l y ) c o r r o b o r a t e d g a i n s r e p o r t e d b y p a r e n t s .', 'H i g h l e v e l s o f p a r e n t , c h i l d a n d s t a f f s a t i s f a c t i o n w e r e r e p o r t e d , a l o n g w i t h h i g h l e v e l s o f t r e a t m e n t f i d e l i t y .', 'S t a n d a r d i z e d e f f e c t s i z e e s t i m a t e s w e r e p r i m a r i l y i n t h e m e d i u m a n d l a r g e r a n g e s a n d f a v o r e d t h e t r e a t m e n t g r o u p .']
model = MODELS['use-char-pos-embed-tribrid']
abstract_pred_probs = model.predict([tf.constant(abstract_lines), tf.constant(abstract_chars),
abstract_line_numbers_one_hot, abstract_total_lines_one_hot])
abstract_pred_probs
array([[0.3974437 , 0.08039878, 0.04826605, 0.41804546, 0.05584609],
[0.10740155, 0.03466441, 0.6462564 , 0.12738277, 0.08429494],
[0.06590997, 0.02248404, 0.7570342 , 0.08790317, 0.06666864],
[0.10426515, 0.1601645 , 0.47555184, 0.07810491, 0.18191367],
[0.04650297, 0.06772824, 0.34206837, 0.04449769, 0.4992027 ],
[0.04270031, 0.05719513, 0.65546656, 0.04226989, 0.20236808],
[0.02312696, 0.08753531, 0.08654262, 0.02455954, 0.7782356 ],
[0.01630326, 0.09151833, 0.12629814, 0.02622827, 0.73965204]],
dtype=float32)
abstract_preds = tf.argmax(abstract_pred_probs, axis=1)
abstract_preds
<tf.Tensor: shape=(8,), dtype=int64, numpy=array([3, 2, 2, 2, 4, 2, 4, 4])>
abstract_pred_class_names = [CLASS_NAMES[i] for i in abstract_preds]
abstract_pred_class_names
['OBJECTIVE', 'METHODS', 'METHODS', 'METHODS', 'RESULTS', 'METHODS', 'RESULTS', 'RESULTS']
for i, line in enumerate(abstract_lines):
print(f'{abstract_pred_class_names[i]}: {line}')
OBJECTIVE: This RCT examined the efficacy of a manualized social intervention for children with HFASDs. METHODS: Participants were randomly assigned to treatment or wait-list conditions. METHODS: Treatment included instruction and therapeutic activities targeting social skills, face-emotion recognition, interest expansion, and interpretation of non-literal language. METHODS: A response-cost program was applied to reduce problem behaviors and foster skills acquisition. RESULTS: Significant treatment effects were found for five of seven primary outcome measures (parent ratings and direct child measures). METHODS: Secondary measures based on staff ratings (treatment group only) corroborated gains reported by parents. RESULTS: High levels of parent, child and staff satisfaction were reported, along with high levels of treatment fidelity. RESULTS: Standardized effect size estimates were primarily in the medium and large ranges and favored the treatment group.